Interordinal chimera formation between medaka and zebrafish for analyzing stem cell differentiation
- PMID: 22204449
- PMCID: PMC3411366
- DOI: 10.1089/scd.2011.0630
Interordinal chimera formation between medaka and zebrafish for analyzing stem cell differentiation
Abstract
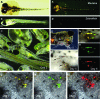
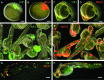
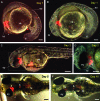
Chimera formation is a standard test for pluripotency of stem cells in vivo. Interspecific chimera formation between distantly related organisms offers also an attractive approach for propagating endangered species. Parameters influencing interspecies chimera formation have remained poorly elucidated. Here, we report interordinal chimera formation between medaka and zebrafish, which separated ∼320 million years ago and exhibit a more than 2-fold difference in developmental speed. We show that, on transplantation into zebrafish blastulae, both noncultivated blastomeres and long-term cultivated embryonic stem (ES) cells of medaka adopted the zebrafish developmental program and differentiated into physiologically functional cell types including pigment cells, blood cells, and cardiomyocytes. We also show that medaka ES cells express differentiation gene markers during chimeric embryogenesis. Therefore, the evolutionary distance and different embryogenesis speeds do not produce donor-host incompatibility to compromise chimera formation between medaka and zebrafish, and molecular markers are valuable for analyzing lineage commitment and cell differentiation in interspecific chimeric embryos.
Figures

Similar articles
-
Medaka insulin-like growth factor-2 supports self-renewal of the embryonic stem cell line and blastomeres in vitro.Sci Rep. 2017 Mar 6;7(1):78. doi: 10.1038/s41598-017-00094-y. Sci Rep. 2017. PMID: 28250437 Free PMC article.
-
Isolation and differentiation of medaka embryonic stem cells.Methods Mol Biol. 2006;329:3-16. doi: 10.1385/1-59745-037-5:3. Methods Mol Biol. 2006. PMID: 16845980
-
Efficiency of cell culture derivation from blastula embryos and of chimera formation in the medaka (Oryzias latipes) depends on donor genotype and passage number.Dev Genes Evol. 1998 Dec;208(10):595-602. doi: 10.1007/s004270050220. Dev Genes Evol. 1998. PMID: 9811979
-
Fish stem cell cultures.Int J Biol Sci. 2011 Apr 13;7(4):392-402. doi: 10.7150/ijbs.7.392. Int J Biol Sci. 2011. PMID: 21547056 Free PMC article. Review.
-
Pigment pattern formation in the medaka embryo.Pigment Cell Res. 2005 Apr;18(2):64-73. doi: 10.1111/j.1600-0749.2005.00216.x. Pigment Cell Res. 2005. PMID: 15760335 Review.
Cited by
-
Dazl is a critical player for primordial germ cell formation in medaka.Sci Rep. 2016 Jun 22;6:28317. doi: 10.1038/srep28317. Sci Rep. 2016. PMID: 27328644 Free PMC article.
-
The Multi-Causal Basis of Developmental Potential Construction.Acta Biotheor. 2023 Jan 30;71(1):6. doi: 10.1007/s10441-023-09456-8. Acta Biotheor. 2023. PMID: 36715846 Free PMC article.
-
Gene transfer and genome-wide insertional mutagenesis by retroviral transduction in fish stem cells.PLoS One. 2015 Jun 1;10(6):e0127961. doi: 10.1371/journal.pone.0127961. eCollection 2015. PLoS One. 2015. PMID: 26029933 Free PMC article.
-
Does resource availability help determine the evolutionary route to multicellularity?Evol Dev. 2019 May;21(3):115-119. doi: 10.1111/ede.12287. Epub 2019 Mar 25. Evol Dev. 2019. PMID: 30912270 Free PMC article.
-
Generation of a Normal Long-Term-Cultured Chinese Hook Snout Carp Spermatogonial Stem Cell Line Capable of Sperm Production In Vitro.Biology (Basel). 2022 Jul 18;11(7):1069. doi: 10.3390/biology11071069. Biology (Basel). 2022. PMID: 36101449 Free PMC article.
References
-
- Evans MJ. Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. - PubMed
-
- Bernemann C. Greber B. Ko K. Sterneckert J. Han DW. Arauzo-Bravo MJ. Scholer HR. Distinct developmental ground states of epiblast stem cell lines determine different pluripotency features. Stem Cells. 2011;29:1496–1503. - PubMed
-
- Reubinoff BE. Pera MF. Fong CY. Trounson A. Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. - PubMed
-
- Hong Y. Winkler C. Schartl M. Pluripotency and differentiation of embryonic stem cell lines from the medakafish (Oryzias latipes) Mech Dev. 1996;60:33–44. - PubMed
-
- Yi M. Hong N. Hong Y. Generation of medaka fish haploid embryonic stem cells. Science. 2009;326:430–433. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

