Heterologous expression of Pycnoporus cinnabarinus cellobiose dehydrogenase in Pichia pastoris and involvement in saccharification processes
- PMID: 22204630
- PMCID: PMC3268779
- DOI: 10.1186/1475-2859-10-113
Heterologous expression of Pycnoporus cinnabarinus cellobiose dehydrogenase in Pichia pastoris and involvement in saccharification processes
Abstract
Background: Cellobiose dehydrogenase (CDH) is an extracellular hemoflavoenzyme produced by lignocellulose-degrading fungi including Pycnoporus cinnabarinus. We investigated the cellulolytic system of P. cinnabarinus, focusing on the involvement of CDH in the deconstruction of lignocellulosic biomass.
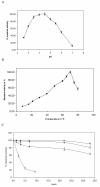
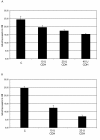
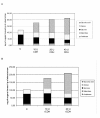
Results: First, P. cinnabarinus growth conditions were optimized for CDH production. Following growth under cellulolytic conditions, the main components secreted were cellulases, xylanases and CDH. To investigate the contribution of P. cinnabarinus secretome in saccharification processes, the Trichoderma reesei enzymatic cocktail was supplemented with the P. cinnabarinus secretome. A significant enhancement of the degradation of wheat straw was observed with (i) the production of a large amount of gluconic acid, (ii) increased hemicellulose degradation, and (iii) increased overall degradation of the lignocellulosic material. P. cinnabarinus CDH was heterologously expressed in Pichia pastoris to obtain large amounts of pure enzyme. In a bioreactor, the recombinant CDH (rCDH) expression level reached 7800 U/L. rCDH exhibited values of biochemical parameters similar to those of the natural enzyme, and was able to bind cellulose despite the absence of a carbohydrate-binding module (CBM). Following supplementation of purified rCDH to T. reesei enzymatic cocktail, formation of gluconic acid and increased hemicellulose degradation were observed, thus confirming the previous results observed with P. cinnabarinus secretome.
Conclusions: We demonstrate that CDH offers an attractive tool for saccharification process enhancement due to gluconic acid production from raw lignocellulosic material.
Figures



Similar articles
-
Production and characterization of recombinant Phanerochaete chrysosporium cellobiose dehydrogenase in the methylotrophic yeast Pichia pastoris.Biosci Biotechnol Biochem. 2001 Sep;65(9):2050-7. doi: 10.1271/bbb.65.2050. Biosci Biotechnol Biochem. 2001. PMID: 11676020
-
Heterologous production of cellobiose dehydrogenases from the basidiomycete Coprinopsis cinerea and the ascomycete Podospora anserina and their effect on saccharification of wheat straw.Appl Microbiol Biotechnol. 2013 Jun;97(11):4873-85. doi: 10.1007/s00253-012-4355-y. Epub 2012 Sep 1. Appl Microbiol Biotechnol. 2013. PMID: 22940800
-
The genome of the white-rot fungus Pycnoporus cinnabarinus: a basidiomycete model with a versatile arsenal for lignocellulosic biomass breakdown.BMC Genomics. 2014 Jun 18;15:486. doi: 10.1186/1471-2164-15-486. BMC Genomics. 2014. PMID: 24942338 Free PMC article.
-
A critical review of cellobiose dehydrogenases.J Biotechnol. 2000 Mar 10;78(2):93-113. doi: 10.1016/s0168-1656(00)00206-6. J Biotechnol. 2000. PMID: 10725534 Review.
-
Methodologies and perspectives of proteomics applied to filamentous fungi: from sample preparation to secretome analysis.Int J Mol Sci. 2015 Mar 12;16(3):5803-29. doi: 10.3390/ijms16035803. Int J Mol Sci. 2015. PMID: 25775160 Free PMC article. Review.
Cited by
-
Scalable methanol-free production of recombinant glucuronoyl esterase in Pichia pastoris.BMC Res Notes. 2019 Sep 18;12(1):596. doi: 10.1186/s13104-019-4638-9. BMC Res Notes. 2019. PMID: 31533815 Free PMC article.
-
Inactivation of Cellobiose Dehydrogenases Modifies the Cellulose Degradation Mechanism of Podospora anserina.Appl Environ Microbiol. 2016 Dec 30;83(2):e02716-16. doi: 10.1128/AEM.02716-16. Print 2017 Jan 15. Appl Environ Microbiol. 2016. PMID: 27836848 Free PMC article.
-
Targeting Penicillium expansum GMC Oxidoreductase with High Affinity Small Molecules for Reducing Patulin Production.Biology (Basel). 2020 Dec 31;10(1):21. doi: 10.3390/biology10010021. Biology (Basel). 2020. PMID: 33396459 Free PMC article.
-
Molecular and catalytic properties of fungal extracellular cellobiose dehydrogenase produced in prokaryotic and eukaryotic expression systems.Microb Cell Fact. 2017 Feb 28;16(1):37. doi: 10.1186/s12934-017-0653-5. Microb Cell Fact. 2017. PMID: 28245812 Free PMC article.
-
Cello-oligosaccharide oxidation reveals differences between two lytic polysaccharide monooxygenases (family GH61) from Podospora anserina.Appl Environ Microbiol. 2013 Jan;79(2):488-96. doi: 10.1128/AEM.02942-12. Epub 2012 Nov 2. Appl Environ Microbiol. 2013. PMID: 23124232 Free PMC article.
References
-
- Martinez D, Larrondo LF, Putnam N, Gelpke MDS, Huang K, Chapman J, Helfenbein KG, Ramaiya P, Detter JC, Larimer F, Coutinho PM, Henrissat B, Berka R, Cullen D, Rokhsar D. Genome sequence of the lignocellulose degrading fungus Phanerochaete chrysosporium strain RP78. Nat Biotechnol. 2004;22:695–700. doi: 10.1038/nbt967. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

