Power tools for gene expression and clonal analysis in Drosophila
- PMID: 22205518
- PMCID: PMC3574576
- DOI: 10.1038/nmeth.1800
Power tools for gene expression and clonal analysis in Drosophila
Abstract
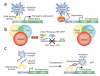
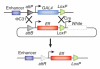
The development of two-component expression systems in Drosophila melanogaster, one of the most powerful genetic models, has allowed the precise manipulation of gene function in specific cell populations. These expression systems, in combination with site-specific recombination approaches, have also led to the development of new methods for clonal lineage analysis. We present a hands-on user guide to the techniques and approaches that have greatly increased resolution of genetic analysis in the fly, with a special focus on their application for lineage analysis. Our intention is to provide guidance and suggestions regarding which genetic tools are most suitable for addressing different developmental questions.
Figures
References
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
-
-
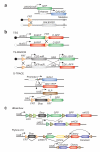
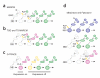
Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–461.. In this important paper for cell-lineage studies, the MARCM technique was described, which allows for the generation of labeled mutant cells in otherwise wild-type organisms.
-
-
-
Griffin R, et al. The twin spot generator for differential Drosophila lineage analysis. Nat. Methods. 2009;6:600–602. Together with reference 6, this paper describes a system for the fruit fly that allows labeling the two progeny of a cell in two different colors, facilitating cell-lineage and gene-function studies.
-
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

