Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits
- PMID: 22205743
- PMCID: PMC3302248
- DOI: 10.1128/JVI.06882-11
Evidence for a common evolutionary origin of coronavirus spike protein receptor-binding subunits
Abstract
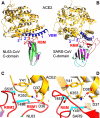
Among different coronavirus genera, the receptor-binding S1 subunits of their spike proteins differ in primary, secondary, and tertiary structures. This study identified shared structural topologies (connectivity of secondary structural elements) in S1 domains of different coronavirus genera. The results suggest that coronavirus S1 subunits share a common evolutionary origin but have attained diverse sequences and structures following extensive divergent evolution. The results also increase understanding of the structures and functions of coronavirus S1 domains whose tertiary structures are currently unknown.
Figures



References
-
- Li F, Li WH, Farzan M, Harrison SC. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864–1868 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

