The Plasmodium falciparum malaria M1 alanyl aminopeptidase (PfA-M1): insights of catalytic mechanism and function from MD simulations
- PMID: 22205955
- PMCID: PMC3244404
- DOI: 10.1371/journal.pone.0028589
The Plasmodium falciparum malaria M1 alanyl aminopeptidase (PfA-M1): insights of catalytic mechanism and function from MD simulations
Abstract
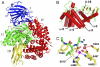
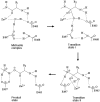
Malaria caused by several species of Plasmodium is major parasitic disease of humans, causing 1-3 million deaths worldwide annually. The widespread resistance of the human parasite to current drug therapies is of major concern making the identification of new drug targets urgent. While the parasite grows and multiplies inside the host erythrocyte it degrades the host cell hemoglobin and utilizes the released amino acids to synthesize its own proteins. The P. falciparum malarial M1 alanyl-aminopeptidase (PfA-M1) is an enzyme involved in the terminal stages of hemoglobin digestion and the generation of an amino acid pool within the parasite. The enzyme has been validated as a potential drug target since inhibitors of the enzyme block parasite growth in vitro and in vivo. In order to gain further understanding of this enzyme, molecular dynamics simulations using data from a recent crystal structure of PfA-M1 were performed. The results elucidate the pentahedral coordination of the catalytic Zn in these metallo-proteases and provide new insights into the roles of this cation and important active site residues in ligand binding and in the hydrolysis of the peptide bond. Based on the data, we propose a two-step catalytic mechanism, in which the conformation of the active site is altered between the Michaelis complex and the transition state. In addition, the simulations identify global changes in the protein in which conformational transitions in the catalytic domain are transmitted at the opening of the N-terminal 8 Å-long channel and at the opening of the 30 Å-long C-terminal internal chamber that facilitates entry of peptides to the active site and exit of released amino acids. The possible implications of these global changes with regard to enzyme function are discussed.
Conflict of interest statement
Figures



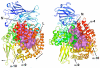
Similar articles
-
KBE009: An antimalarial bestatin-like inhibitor of the Plasmodium falciparum M1 aminopeptidase discovered in an Ugi multicomponent reaction-derived peptidomimetic library.Bioorg Med Chem. 2017 Sep 1;25(17):4628-4636. doi: 10.1016/j.bmc.2017.06.047. Epub 2017 Jul 4. Bioorg Med Chem. 2017. PMID: 28728898
-
Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase.Proc Natl Acad Sci U S A. 2009 Feb 24;106(8):2537-42. doi: 10.1073/pnas.0807398106. Epub 2009 Feb 5. Proc Natl Acad Sci U S A. 2009. PMID: 19196988 Free PMC article.
-
Mapping the Pathway and Dynamics of Bestatin Inhibition of the Plasmodium falciparum M1 Aminopeptidase PfA-M1.ChemMedChem. 2018 Dec 6;13(23):2504-2513. doi: 10.1002/cmdc.201800563. Epub 2018 Nov 9. ChemMedChem. 2018. PMID: 30318749
-
Plasmodium falciparum neutral aminopeptidases: new targets for anti-malarials.Trends Biochem Sci. 2010 Jan;35(1):53-61. doi: 10.1016/j.tibs.2009.08.004. Epub 2009 Sep 30. Trends Biochem Sci. 2010. PMID: 19796954 Review.
-
Plasmodium falciparum M1-aminopeptidase: a promising target for the development of antimalarials.Curr Drug Targets. 2014;15(12):1144-65. doi: 10.2174/1389450115666141024115641. Curr Drug Targets. 2014. PMID: 25341419 Review.
Cited by
-
Steered molecular dynamics simulations reveal critical residues for (un)binding of substrates, inhibitors and a product to the malarial M1 aminopeptidase.PLoS Comput Biol. 2018 Oct 31;14(10):e1006525. doi: 10.1371/journal.pcbi.1006525. eCollection 2018 Oct. PLoS Comput Biol. 2018. PMID: 30379805 Free PMC article.
-
Driving antimalarial design through understanding of target mechanism.Biochem Soc Trans. 2020 Oct 30;48(5):2067-2078. doi: 10.1042/BST20200224. Biochem Soc Trans. 2020. PMID: 32869828 Free PMC article. Review.
-
On-target, dual aminopeptidase inhibition provides cross-species antimalarial activity.mBio. 2024 Jun 12;15(6):e0096624. doi: 10.1128/mbio.00966-24. Epub 2024 May 8. mBio. 2024. PMID: 38717141 Free PMC article.
-
The X-ray crystal structure of human aminopeptidase N reveals a novel dimer and the basis for peptide processing.J Biol Chem. 2012 Oct 26;287(44):36804-13. doi: 10.1074/jbc.M112.398842. Epub 2012 Aug 29. J Biol Chem. 2012. PMID: 22932899 Free PMC article.
-
Dermal fibroblast cells interactions with single and triple bacterial-species biofilms.Mol Biol Rep. 2021 Apr;48(4):3393-3404. doi: 10.1007/s11033-021-06391-0. Epub 2021 May 19. Mol Biol Rep. 2021. PMID: 34009564
References
-
- Rosenthal PJ. Antimalarial drug discovery: old and new approaches. J Exp Biol. 2003;206:3735–3744. - PubMed
-
- Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, et al. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. - PubMed
-
- Schlitzer M. Malaria chemotherapeutics part I: History of antimalarial drug development, currently used therapeutics, and drugs in clinical development. ChemMedChem. 2007;2:944–986. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

