Clag9 is not essential for PfEMP1 surface expression in non-cytoadherent Plasmodium falciparum parasites with a chromosome 9 deletion
- PMID: 22205992
- PMCID: PMC3242772
- DOI: 10.1371/journal.pone.0029039
Clag9 is not essential for PfEMP1 surface expression in non-cytoadherent Plasmodium falciparum parasites with a chromosome 9 deletion
Abstract
Background: The expression of the clonally variant virulence factor PfEMP1 mediates the sequestration of Plasmodium falciparum infected erythrocytes in the host vasculature and contributes to chronic infection. Non-cytoadherent parasites with a chromosome 9 deletion lack clag9, a gene linked to cytoadhesion in previous studies. Here we present new clag9 data that challenge this view and show that surface the non-cytoadherence phenotype is linked to the expression of a non-functional PfEMP1.
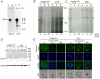
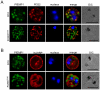
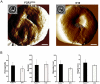
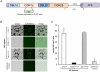
Methodology/principal findings: Loss of adhesion in P. falciparum D10, a parasite line with a large chromosome 9 deletion, was investigated. Surface iodination analysis of non-cytoadherent D10 parasites and COS-7 surface expression of the CD36-binding PfEMP1 CIDR1α domain were performed and showed that these parasites express an unusual trypsin-resistant, non-functional PfEMP1 at the erythrocyte surface. However, the CIDR1α domain of this var gene expressed in COS-7 cells showed strong binding to CD36. Atomic Force Microscopy showed a slightly modified D10 knob morphology compared to adherent parasites. Trafficking of PfEMP1 and KAHRP remained functional in D10. We link the non-cytoadherence phenotype to a chromosome 9 breakage and healing event resulting in the loss of 25 subtelomeric genes including clag9. In contrast to previous studies, knockout of the clag9 gene from 3D7 did not interfere with parasite adhesion to CD36.
Conclusions/significance: Our data show the surface expression of non-functional PfEMP1 in D10 strongly indicating that genes other than clag9 deleted from chromosome 9 are involved in this virulence process possibly via post-translational modifications.
Conflict of interest statement
Figures


Similar articles
-
Targeted disruption of a ring-infected erythrocyte surface antigen (RESA)-like export protein gene in Plasmodium falciparum confers stable chondroitin 4-sulfate cytoadherence capacity.J Biol Chem. 2014 Dec 5;289(49):34408-21. doi: 10.1074/jbc.M114.615393. Epub 2014 Oct 23. J Biol Chem. 2014. PMID: 25342752 Free PMC article.
-
Dual stage synthesis and crucial role of cytoadherence-linked asexual gene 9 in the surface expression of malaria parasite var proteins.Proc Natl Acad Sci U S A. 2010 Sep 21;107(38):16643-8. doi: 10.1073/pnas.1002568107. Epub 2010 Sep 7. Proc Natl Acad Sci U S A. 2010. PMID: 20823248 Free PMC article.
-
Discovery of a novel and conserved Plasmodium falciparum exported protein that is important for adhesion of PfEMP1 at the surface of infected erythrocytes.Cell Microbiol. 2015 Aug;17(8):1205-16. doi: 10.1111/cmi.12430. Epub 2015 Mar 16. Cell Microbiol. 2015. PMID: 25703704 Free PMC article.
-
The role of PfEMP1 adhesion domain classification in Plasmodium falciparum pathogenesis research.Mol Biochem Parasitol. 2014 Jul;195(2):82-7. doi: 10.1016/j.molbiopara.2014.07.006. Epub 2014 Jul 23. Mol Biochem Parasitol. 2014. PMID: 25064606 Free PMC article. Review.
-
Malaria's deadly grip: cytoadhesion of Plasmodium falciparum-infected erythrocytes.Cell Microbiol. 2013 Dec;15(12):1976-83. doi: 10.1111/cmi.12183. Epub 2013 Sep 4. Cell Microbiol. 2013. PMID: 23957661 Free PMC article. Review.
Cited by
-
The Plasmodium falciparum rhoptry protein RhopH3 plays essential roles in host cell invasion and nutrient uptake.Elife. 2017 Mar 2;6:e23239. doi: 10.7554/eLife.23239. Elife. 2017. PMID: 28252384 Free PMC article.
-
How Malaria Parasites Acquire Nutrients From Their Host.Front Cell Dev Biol. 2021 Mar 25;9:649184. doi: 10.3389/fcell.2021.649184. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33842474 Free PMC article. Review.
-
Plasmodium falciparum CLAG Paralogs All Traffic to the Host Membrane but Knockouts Have Distinct Phenotypes.Microorganisms. 2024 Jun 8;12(6):1172. doi: 10.3390/microorganisms12061172. Microorganisms. 2024. PMID: 38930554 Free PMC article.
-
The conserved clag multigene family of malaria parasites: essential roles in host-pathogen interaction.Drug Resist Updat. 2015 Jan;18:47-54. doi: 10.1016/j.drup.2014.10.004. Epub 2014 Nov 3. Drug Resist Updat. 2015. PMID: 25467627 Free PMC article. Review.
-
Schizont transcriptome variation among clinical isolates and laboratory-adapted clones of the malaria parasite Plasmodium falciparum.BMC Genomics. 2018 Dec 10;19(1):894. doi: 10.1186/s12864-018-5257-x. BMC Genomics. 2018. PMID: 30526479 Free PMC article.
References
-
- Kraemer SM, Smith JD. A family affair: var genes, PfEMP1 binding, and malaria disease. Curr Opin Microbiol. 2006;9:374–380. - PubMed
-
- Rasti N, Wahlgren M, Chen Q. Molecular aspects of malaria pathogenesis. FEMS Immunol Med Microbiol. 2004;41:9–26. - PubMed
-
- Maier AG, Cooke BM, Cowman AF, Tilley L. Malaria parasite proteins that remodel the host erythrocyte. Nat Rev Microbiol. 2009;7:341–354. - PubMed
-
- Baruch DI, Ma XC, Singh HB, Bi X, Pasloske BL, et al. Identification of a region of PfEMP1 that mediates adherence of Plasmodium falciparum infected erythrocytes to CD36: conserved function with variant sequence. Blood. 1997;90:3766–3775. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

