Architecture and conservation of the bacterial DNA replication machinery, an underexploited drug target
- PMID: 22206257
- PMCID: PMC3290774
- DOI: 10.2174/138945012799424598
Architecture and conservation of the bacterial DNA replication machinery, an underexploited drug target
Abstract
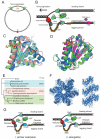
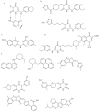
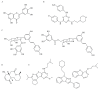
New antibiotics with novel modes of action are required to combat the growing threat posed by multi-drug resistant bacteria. Over the last decade, genome sequencing and other high-throughput techniques have provided tremendous insight into the molecular processes underlying cellular functions in a wide range of bacterial species. We can now use these data to assess the degree of conservation of certain aspects of bacterial physiology, to help choose the best cellular targets for development of new broad-spectrum antibacterials. DNA replication is a conserved and essential process, and the large number of proteins that interact to replicate DNA in bacteria are distinct from those in eukaryotes and archaea; yet none of the antibiotics in current clinical use acts directly on the replication machinery. Bacterial DNA synthesis thus appears to be an underexploited drug target. However, before this system can be targeted for drug design, it is important to understand which parts are conserved and which are not, as this will have implications for the spectrum of activity of any new inhibitors against bacterial species, as well as the potential for development of drug resistance. In this review we assess similarities and differences in replication components and mechanisms across the bacteria, highlight current progress towards the discovery of novel replication inhibitors, and suggest those aspects of the replication machinery that have the greatest potential as drug targets.
Figures
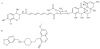
Similar articles
-
Bacterial DNA replication enzymes as targets for antibacterial drug discovery.Expert Opin Drug Discov. 2012 Apr;7(4):327-39. doi: 10.1517/17460441.2012.660478. Epub 2012 Feb 21. Expert Opin Drug Discov. 2012. PMID: 22458504 Review.
-
Targeting bacterial RNA polymerase σ70 for development of broad-spectrum antisense antibacterials.Recent Pat Antiinfect Drug Discov. 2012 Dec 1;7(3):213-22. doi: 10.2174/157489112803521986. Recent Pat Antiinfect Drug Discov. 2012. PMID: 22742395 Review.
-
DNA replication proteins as potential targets for antimicrobials in drug-resistant bacterial pathogens.J Antimicrob Chemother. 2017 May 1;72(5):1275-1284. doi: 10.1093/jac/dkw548. J Antimicrob Chemother. 2017. PMID: 28073967 Free PMC article. Review.
-
DNA replication in the third domain (of life).Curr Protein Pept Sci. 2000 Sep;1(2):139-54. doi: 10.2174/1389203003381414. Curr Protein Pept Sci. 2000. PMID: 12369914 Review.
-
Toward Understanding the Chemistry and Biology of 1-Deoxy-d-xylulose 5-Phosphate (DXP) Synthase: A Unique Antimicrobial Target at the Heart of Bacterial Metabolism.Acc Chem Res. 2018 Oct 16;51(10):2546-2555. doi: 10.1021/acs.accounts.8b00321. Epub 2018 Sep 11. Acc Chem Res. 2018. PMID: 30203647 Free PMC article.
Cited by
-
Rapid single-molecule characterisation of enzymes involved in nucleic-acid metabolism.Nucleic Acids Res. 2023 Jan 11;51(1):e5. doi: 10.1093/nar/gkac949. Nucleic Acids Res. 2023. PMID: 36321650 Free PMC article.
-
Protein-protein interactions in bacteria: a promising and challenging avenue towards the discovery of new antibiotics.Beilstein J Org Chem. 2018 Nov 21;14:2881-2896. doi: 10.3762/bjoc.14.267. eCollection 2018. Beilstein J Org Chem. 2018. PMID: 30546472 Free PMC article. Review.
-
Fragment-Based Discovery of Inhibitors of the Bacterial DnaG-SSB Interaction.Antibiotics (Basel). 2018 Feb 22;7(1):14. doi: 10.3390/antibiotics7010014. Antibiotics (Basel). 2018. PMID: 29470422 Free PMC article.
-
Comprehensive analysis of DNA polymerase III α subunits and their homologs in bacterial genomes.Nucleic Acids Res. 2014 Feb;42(3):1393-413. doi: 10.1093/nar/gkt900. Epub 2013 Oct 7. Nucleic Acids Res. 2014. PMID: 24106089 Free PMC article.
-
Primase is required for helicase activity and helicase alters the specificity of primase in the enteropathogen Clostridium difficile.Open Biol. 2016 Dec;6(12):160272. doi: 10.1098/rsob.160272. Open Biol. 2016. PMID: 28003473 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
