A two-dimensional ERK-AKT signaling code for an NGF-triggered cell-fate decision
- PMID: 22206868
- PMCID: PMC3897208
- DOI: 10.1016/j.molcel.2011.11.023
A two-dimensional ERK-AKT signaling code for an NGF-triggered cell-fate decision
Abstract
Growth factors activate Ras, PI3K, and other signaling pathways. It is not well understood how these signals are translated by individual cells into a decision to proliferate or differentiate. Here, using single-cell image analysis of nerve growth factor (NGF)-stimulated PC12 cells, we identified a two-dimensional phospho-ERK (pERK)-phospho-AKT (pAKT) response map with a curved boundary that separates differentiating from proliferating cells. The boundary position remained invariant when different stimuli were used or upstream signaling components perturbed. We further identified Rasa2 as a negative feedback regulator that links PI3K to Ras, placing the stochastically distributed pERK-pAKT signals close to the decision boundary. This allows for uniform NGF stimuli to create a subpopulation of cells that differentiates with each cycle of proliferation. Thus, by linking a complex signaling system to a simpler intermediate response map, cells gain unique integration and control capabilities to balance cell number expansion with differentiation.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
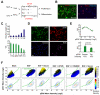
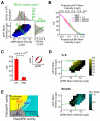
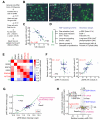
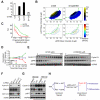
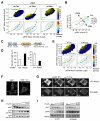
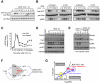
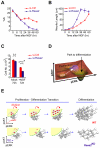
Comment in
-
Cellular signal processing: out of one, many.Mol Cell. 2012 Jan 27;45(2):143-4. doi: 10.1016/j.molcel.2012.01.004. Mol Cell. 2012. PMID: 22284673
References
-
- Albert R. Scale-free networks in cell biology. Journal of cell science. 2005;118:4947–4957. - PubMed
-
- Arias AM, Hayward P. Filtering transcriptional noise during development: concepts and mechanisms. Nat Rev Genet. 2006;7:34–44. - PubMed
-
- Bar-Sagi D, Feramisco JR. Microinjection of the ras oncogene protein into PC12 cells induces morphological differentiation. Cell. 1985;42:841–848. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

