Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice
- PMID: 22207572
- PMCID: PMC3269871
- DOI: 10.1105/tpc.111.090050
Positive autoregulation of a KNOX gene is essential for shoot apical meristem maintenance in rice
Abstract
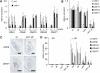
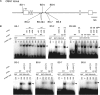
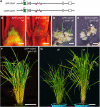
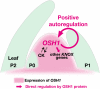
Self-maintenance of the shoot apical meristem (SAM), from which aerial organs are formed throughout the life cycle, is crucial in plant development. Class I Knotted1-like homeobox (KNOX) genes restrict cell differentiation and play an indispensable role in maintaining the SAM. However, the mechanism that positively regulates their expression is unknown. Here, we show that expression of a rice (Oryza sativa) KNOX gene, Oryza sativa homeobox1 (OSH1), is positively regulated by direct autoregulation. Interestingly, loss-of-function mutants of OSH1 lose the SAM just after germination but can be rescued to grow until reproductive development when they are regenerated from callus. Double mutants of osh1 and d6, a loss-of-function mutant of OSH15, fail to establish the SAM both in embryogenesis and regeneration. Expression analyses in these mutants reveal that KNOX gene expression is positively regulated by the phytohormone cytokinin and by KNOX genes themselves. We demonstrate that OSH1 directly binds to five KNOX loci, including OSH1 and OSH15, through evolutionarily conserved cis-elements and that the positive autoregulation of OSH1 is indispensable for its own expression and SAM maintenance. Thus, the maintenance of the indeterminate state mediated by positive autoregulation of a KNOX gene is an indispensable mechanism of self-maintenance of the SAM.
Figures




References
-
- Byrne M.E., Barley R., Curtis M., Arroyo J.M., Dunham M., Hudson A., Martienssen R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408: 967–971 - PubMed
-
- Byrne M.E., Simorowski J., Martienssen R.A. (2002). ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129: 1957–1965 - PubMed
-
- Chiu W., Niwa Y., Zeng W., Hirano T., Kobayashi H., Sheen J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6: 325–330 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

