Transcript and protein profiling analysis of OTA-induced cell death reveals the regulation of the toxicity response process in Arabidopsis thaliana
- PMID: 22207617
- PMCID: PMC3295405
- DOI: 10.1093/jxb/err447
Transcript and protein profiling analysis of OTA-induced cell death reveals the regulation of the toxicity response process in Arabidopsis thaliana
Abstract
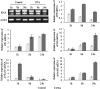
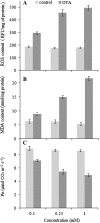
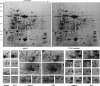
Ochratoxin A (OTA) is a toxic isocoumarin derivative produced by various species of mould which mainly grow on grain, coffee, and nuts. Recent studies have suggested that OTA induces cell death in plants. To investigate possible mechanisms of OTA phytotoxicity, both digital gene expression (DGE) transcriptomic and two-dimensional electrophoresis proteomic analyses were used, through which 3118 genes and 23 proteins were identified as being up- or down-regulated at least 2-fold in Arabidopsis leaf in response to OTA treatment. First, exposure of excised Arabidopsis thaliana leaves to OTA rapidly causes the hypersensitive reponse, significantly accelerates the increase of reactive oxygen species and malondialdehyde, and enhances antioxidant enzyme defence responses and xenobiotic detoxification. Secondly, OTA stimulation causes dynamic changes in transcription factors and activates the membrane transport system dramatically. Thirdly, a concomitant persistence of compromised photosynthesis and photorespiration is indicative of a metabolic shift from a highly active to a weak state. Finally, the data revealed that ethylene, salicylic acid, jasmonic acid, and mitogen-activated protein kinase signalling molecules mediate the process of toxicity caused by OTA. Profiling analyses on Arabidopsis in response to OTA will provide new insights into signalling transduction that modulates the OTA phytotoxicity mechanism, facilitate mapping of regulatory networks, and extend the ability to improve OTA tolerance in Arabidopsis.
Figures





References
-
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Research. 1997;7:986–995. - PubMed
-
- Bednarek P, Piślewska-Bednarek M, Svatoš A, et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323:101–106. - PubMed
-
- Boesch-Saadatmandi C, Loboda A, Jozkowicz A, Huebbe P, Blank R, Wolffram S, Dulak J, Rimbach G. Effect of ochratoxin A on redox-regulated transcription factors, antioxidant enzymes and glutathione-S-transferase in cultured kidney tubulus cells. Food and Chemical Toxicology. 2008;46:2665–2671. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

