Human cancer cells express Slug-based epithelial-mesenchymal transition gene expression signature obtained in vivo
- PMID: 22208948
- PMCID: PMC3268117
- DOI: 10.1186/1471-2407-11-529
Human cancer cells express Slug-based epithelial-mesenchymal transition gene expression signature obtained in vivo
Abstract
Background: The biological mechanisms underlying cancer cell motility and invasiveness remain unclear, although it has been hypothesized that they involve some type of epithelial-mesenchymal transition (EMT).
Methods: We used xenograft models of human cancer cells in immunocompromised mice, profiling the harvested tumors separately with species-specific probes and computationally analyzing the results.
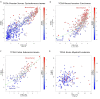
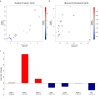
Results: Here we show that human cancer cells express in vivo a precise multi-cancer invasion-associated gene expression signature that prominently includes many EMT markers, among them the transcription factor Slug, fibronectin, and α-SMA. We found that human, but not mouse, cells express the signature and Slug is the only upregulated EMT-inducing transcription factor. The signature is also present in samples from many publicly available cancer gene expression datasets, suggesting that it is produced by the cancer cells themselves in multiple cancer types, including nonepithelial cancers such as neuroblastoma. Furthermore, we found that the presence of the signature in human xenografted cells was associated with a downregulation of adipocyte markers in the mouse tissue adjacent to the invasive tumor, suggesting that the signature is triggered by contextual microenvironmental interactions when the cancer cells encounter adipocytes, as previously reported.
Conclusions: The known, precise and consistent gene composition of this cancer mesenchymal transition signature, particularly when combined with simultaneous analysis of the adjacent microenvironment, provides unique opportunities for shedding light on the underlying mechanisms of cancer invasiveness as well as identifying potential diagnostic markers and targets for metastasis-inhibiting therapeutics.
Figures
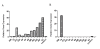

Comment in
-
An interplay between epithelial: mesenchymal transition and reverse: gene patterns for tumorigenesis.Biomark Med. 2012 Apr;6(2):199-200. doi: 10.2217/BMM.12.5. Biomark Med. 2012. PMID: 22550754 No abstract available.
References
-
- Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, Hartwell K, Onder TT, Gupta PB, Evans KW. et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107(35):15449–15454. doi: 10.1073/pnas.1004900107. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

