DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy
- PMID: 22209328
- PMCID: PMC3264808
- DOI: 10.1016/j.devcel.2011.11.013
DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy
Abstract
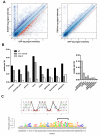
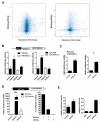
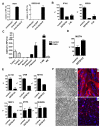
Facioscapulohumeral dystrophy (FSHD) is one of the most common inherited muscular dystrophies. The causative gene remains controversial and the mechanism of pathophysiology unknown. Here we identify genes associated with germline and early stem cell development as targets of the DUX4 transcription factor, a leading candidate gene for FSHD. The genes regulated by DUX4 are reliably detected in FSHD muscle but not in controls, providing direct support for the model that misexpression of DUX4 is a causal factor for FSHD. Additionally, we show that DUX4 binds and activates LTR elements from a class of MaLR endogenous primate retrotransposons and suppresses the innate immune response to viral infection, at least in part through the activation of DEFB103, a human defensin that can inhibit muscle differentiation. These findings suggest specific mechanisms of FSHD pathology and identify candidate biomarkers for disease diagnosis and progression.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


References
-
- Booth HA, Holland PW. Annotation, nomenclature and evolution of four novel homeobox genes expressed in the human germ line. Gene. 2007;387:7–14. - PubMed
-
- Bosch N, Caceres M, Cardone MF, Carreras A, Ballana E, Rocchi M, Armengol L, Estivill X. Characterization and evolution of the novel gene family FAM90A in primates originated by multiple duplication and rearrangement events. Hum Mol Genet. 2007;16:2572–2582. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

