Going in the right direction: mating-type switching of Schizosaccharomyces pombe is controlled by judicious expression of two different swi2 transcripts
- PMID: 22209903
- PMCID: PMC3296259
- DOI: 10.1534/genetics.111.137109
Going in the right direction: mating-type switching of Schizosaccharomyces pombe is controlled by judicious expression of two different swi2 transcripts
Abstract
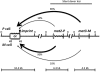
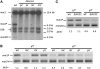
Schizosaccharomyces pombe, the fission yeast, cells alternate between P- and M-mating type, controlled by the alternate alleles of the mating-type locus (mat1). The mat1 switching occurs by replacing mat1 with a copy derived from a silenced "donor locus," mat2P or mat3M. The mechanism of donor choice ensuring that switching occurs primarily and productively to the opposite type, called directionality, is largely unknown. Here we identified the mat1-Mc gene, a mammalian sex-determination gene (SRY) homolog, as the primary gene that dictates directionality in M cells. A previously unrecognized, shorter swi2 mRNA, a truncated form of the swi2, was identified, and its expression requires the mat1-Mc function. We also found that the abp1 gene (human CENPB homolog) controls directionality through swi2 regulation. In addition, we implicated a cis-acting DNA sequence in mat2 utilization. Overall, we showed that switching directionality is controlled by judicious expression of two swi2 transcripts through a cell-type-regulated dual promoter. In this respect, this regulation mechanism resembles that of the Drosophila sex-determination Slx gene.
Figures





References
-
- Arcangioli B., Thon G., 2003. Mating-types cassettes: structure, switching and silencing, pp. 129–147 Molecular Biology of Schizosaccharomyces pombe, edited by Egel R. Springer Verlag, Berlin
-
- Beach D. H., 1983. Cell type switching by DNA transposition in fission yeast. Nature 305: 682–688
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

