Intraspecies biodiversity of the genetically homologous species Brucella microti
- PMID: 22210211
- PMCID: PMC3294491
- DOI: 10.1128/AEM.06351-11
Intraspecies biodiversity of the genetically homologous species Brucella microti
Abstract
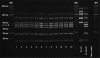
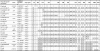
Brucellosis is one of the major bacterial zoonoses worldwide. In the past decade, an increasing number of atypical Brucella strains and species have been described. Brucella microti in particular has attracted attention, because this species not only infects mammalian hosts but also persists in soil. An environmental reservoir may pose a new public health risk, leading to the reemergence of brucellosis. In a polyphasic approach, comprising conventional microbiological techniques and extensive biochemical and molecular techniques, all currently available Brucella microti strains were characterized. While differing in their natural habitats and host preferences, B. microti isolates were found to possess identical 16S rRNA, recA, omp2a, and omp2b gene sequences and identical multilocus sequence analysis (MLSA) profiles at 21 different genomic loci. Only highly variable microsatellite markers of multiple-locus variable-number tandem repeat (VNTR) analysis comprising 16 loci (MLVA-16) showed intraspecies discriminatory power. In contrast, biotyping demonstrated striking differences within the genetically homologous species. The majority of the mammalian isolates agglutinated only with monospecific anti-M serum, whereas soil isolates agglutinated with anti-A, anti-M, and anti-R sera. Bacteria isolated from animal sources were lysed by phages F1, F25, Tb, BK2, Iz, and Wb, whereas soil isolates usually were not. Rough strains of environmental origin were lysed only by phage R/C. B. microti exhibited high metabolic activities similar to those of closely related soil organisms, such as Ochrobactrum spp. Each strain was tested with 93 different substrates and showed an individual metabolic profile. In summary, the adaptation of Brucella microti to a specific habitat or host seems to be a matter of gene regulation rather than a matter of gene configuration.
Figures


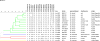

References
-
- Al Dahouk S, et al. 2007. Evaluation of Brucella MLVA typing for human brucellosis. J. Microbiol. Methods 69:137–145 - PubMed
-
- Al Dahouk S, et al. 2005. Seroprevalence of brucellosis, tularemia, and yersiniosis in wild boars (Sus scrofa) from north-eastern Germany. J. Vet. Med. B Infect. Dis. Vet. Public Health 52:444–455 - PubMed
-
- Alton GG, Jones LM, Angus RD, Verger JM. 1988. Techniques for the brucellosis laboratory. Institut National de la Recherche Agronomique, Paris, France
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

