Plasmid localization and organization of melamine degradation genes in Rhodococcus sp. strain Mel
- PMID: 22210223
- PMCID: PMC3294463
- DOI: 10.1128/AEM.06468-11
Plasmid localization and organization of melamine degradation genes in Rhodococcus sp. strain Mel
Abstract
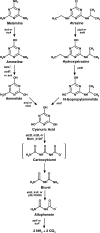
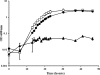
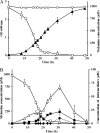
Rhodococcus sp. strain Mel was isolated from soil by enrichment and grew in minimal medium with melamine as the sole N source with a doubling time of 3.5 h. Stoichiometry studies showed that all six nitrogen atoms of melamine were assimilated. The genome was sequenced by Roche 454 pyrosequencing to 13× coverage, and a 22.3-kb DNA region was found to contain a homolog to the melamine deaminase gene trzA. Mutagenesis studies showed that the cyanuric acid hydrolase and biuret hydrolase genes were clustered together on a different 17.9-kb contig. Curing and gene transfer studies indicated that 4 of 6 genes required for the complete degradation of melamine were located on an ∼265-kb self-transmissible linear plasmid (pMel2), but this plasmid was not required for ammeline deamination. The Rhodococcus sp. strain Mel melamine metabolic pathway genes were located in at least three noncontiguous regions of the genome, and the plasmid-borne genes encoding enzymes for melamine metabolism were likely recently acquired.
Figures
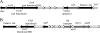

Similar articles
-
Mineralization of melamine and cyanuric acid as sole nitrogen source by newly isolated Arthrobacter spp. using a soil-charcoal perfusion method.World J Microbiol Biotechnol. 2015 May;31(5):785-93. doi: 10.1007/s11274-015-1832-3. Epub 2015 Mar 10. World J Microbiol Biotechnol. 2015. PMID: 25752233
-
Genome sequence of Rhodococcus sp. strain R04, a polychlorinated-biphenyl biodegrader.J Bacteriol. 2011 Sep;193(18):5032-3. doi: 10.1128/JB.05635-11. Epub 2011 Jul 8. J Bacteriol. 2011. PMID: 21742862 Free PMC article.
-
Biodegradation of dioxin by a newly isolated Rhodococcus sp. with the involvement of self-transmissible plasmids.Appl Microbiol Biotechnol. 2013 Jun;97(12):5585-95. doi: 10.1007/s00253-012-4363-y. Epub 2012 Sep 1. Appl Microbiol Biotechnol. 2013. PMID: 22940801
-
Comparative Genomics and Metabolic Analysis Reveals Peculiar Characteristics of Rhodococcus opacus Strain M213 Particularly for Naphthalene Degradation.PLoS One. 2016 Aug 17;11(8):e0161032. doi: 10.1371/journal.pone.0161032. eCollection 2016. PLoS One. 2016. PMID: 27532207 Free PMC article.
-
Biotechnological Potential of Rhodococcus Biodegradative Pathways.J Microbiol Biotechnol. 2018 Jul 28;28(7):1037-1051. doi: 10.4014/jmb.1712.12017. J Microbiol Biotechnol. 2018. PMID: 29913546 Review.
Cited by
-
Cyanuric Acid Biodegradation via Biuret: Physiology, Taxonomy, and Geospatial Distribution.Appl Environ Microbiol. 2020 Jan 7;86(2):e01964-19. doi: 10.1128/AEM.01964-19. Print 2020 Jan 7. Appl Environ Microbiol. 2020. PMID: 31676480 Free PMC article.
-
Mineralization of melamine and cyanuric acid as sole nitrogen source by newly isolated Arthrobacter spp. using a soil-charcoal perfusion method.World J Microbiol Biotechnol. 2015 May;31(5):785-93. doi: 10.1007/s11274-015-1832-3. Epub 2015 Mar 10. World J Microbiol Biotechnol. 2015. PMID: 25752233
-
Ancient Evolution and Recent Evolution Converge for the Biodegradation of Cyanuric Acid and Related Triazines.Appl Environ Microbiol. 2016 Jan 4;82(6):1638-1645. doi: 10.1128/AEM.03594-15. Appl Environ Microbiol. 2016. PMID: 26729715 Free PMC article. Review.
-
Expanding the cyanuric acid hydrolase protein family to the fungal kingdom.J Bacteriol. 2013 Dec;195(23):5233-41. doi: 10.1128/JB.00965-13. Epub 2013 Sep 13. J Bacteriol. 2013. PMID: 24039269 Free PMC article.
-
Microbial changes linked to the accelerated degradation of the herbicide atrazine in a range of temperate soils.Environ Sci Pollut Res Int. 2017 Mar;24(8):7359-7374. doi: 10.1007/s11356-017-8377-y. Epub 2017 Jan 20. Environ Sci Pollut Res Int. 2017. PMID: 28108915 Free PMC article.
References
-
- Assaf NA, Dick WA. 1993. Spheroplast formation and plasmid isolation from Rhodococcus spp. Biotechniques 15:1010–1012 - PubMed
-
- Bruno WJ, Socci ND, Halpern AL. 2000. Weighted neighbor joining: a likelihood-based approach to distance-based phylogeny reconstruction. Mol. Biol. Evol. 17:189–197 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

