Insights from the architecture of the bacterial transcription apparatus
- PMID: 22210308
- PMCID: PMC3769190
- DOI: 10.1016/j.jsb.2011.12.013
Insights from the architecture of the bacterial transcription apparatus
Abstract
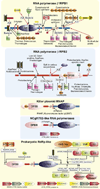
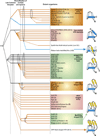
We provide a portrait of the bacterial transcription apparatus in light of the data emerging from structural studies, sequence analysis and comparative genomics to bring out important but underappreciated features. We first describe the key structural highlights and evolutionary implications emerging from comparison of the cellular RNA polymerase subunits with the RNA-dependent RNA polymerase involved in RNAi in eukaryotes and their homologs from newly identified bacterial selfish elements. We describe some previously unnoticed domains and the possible evolutionary stages leading to the RNA polymerases of extant life forms. We then present the case for the ancient orthology of the basal transcription factors, the sigma factor and TFIIB, in the bacterial and the archaeo-eukaryotic lineages. We also present a synopsis of the structural and architectural taxonomy of specific transcription factors and their genome-scale demography. In this context, we present certain notable deviations from the otherwise invariant proteome-wide trends in transcription factor distribution and use it to predict the presence of an unusual lineage-specifically expanded signaling system in certain firmicutes like Paenibacillus. We then discuss the intersection between functional properties of transcription factors and the organization of transcriptional networks. Finally, we present some of the interesting evolutionary conundrums posed by our newly gained understanding of the bacterial transcription apparatus and potential areas for future explorations.
Published by Elsevier Inc.
Figures
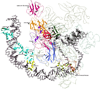



References
-
- Ammelburg M, Frickey T, Lupas AN. Classification of AAA+ proteins. J. Struct. Biol. 2006;156:2–11. - PubMed
-
- Anantharaman V, Aravind L. Diversification of catalytic activities and ligand interactions in the protein fold shared by the sugar isomerases, eIF2B, DeoR transcription factors, acyl-CoA transferases and methenyltetrahydrofolate synthetase. J. Mol. Biol. 2006;356:823–842. - PubMed
-
- Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol. 2001;307:1271–1292. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

