Local unfolding of Cu, Zn superoxide dismutase monomer determines the morphology of fibrillar aggregates
- PMID: 22210350
- PMCID: PMC3320695
- DOI: 10.1016/j.jmb.2011.12.029
Local unfolding of Cu, Zn superoxide dismutase monomer determines the morphology of fibrillar aggregates
Abstract
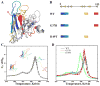
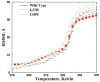
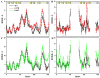
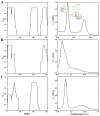
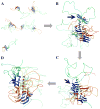
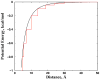
Aggregation of Cu, Zn superoxide dismutase (SOD1) is often found in amyotrophic lateral sclerosis patients. The fibrillar aggregates formed by wild type and various disease-associated mutants have recently been found to have distinct cores and morphologies. Previous computational and experimental studies of wild-type SOD1 suggest that the apo-monomer, highly aggregation prone, displays substantial local unfolding dynamics. The residual folded structure of locally unfolded apoSOD1 corresponds to peptide segments forming the aggregation core as identified by a combination of proteolysis and mass spectroscopy. Therefore, we hypothesize that the destabilization of apoSOD1 caused by various mutations leads to distinct local unfolding dynamics. The partially unfolded structure, exposing the hydrophobic core and backbone hydrogen bond donors and acceptors, is prone to aggregate. The peptide segments in the residual folded structures form the "building block" for aggregation, which in turn determines the morphology of the aggregates. To test this hypothesis, we apply a multiscale simulation approach to study the aggregation of three typical SOD1 variants: wild type, G37R, and I149T. Each of these SOD1 variants has distinct peptide segments forming the core structure and features different aggregate morphologies. We perform atomistic molecular dynamics simulations to study the conformational dynamics of apoSOD1 monomer and coarse-grained molecular dynamics simulations to study the aggregation of partially unfolded SOD1 monomers. Our computational studies of monomer local unfolding and the aggregation of different SOD1 variants are consistent with experiments, supporting the hypothesis of the formation of aggregation "building blocks" via apo-monomer local unfolding as the mechanism of SOD1 fibrillar aggregation.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures

Similar articles
-
Solvent sensitivity of protein aggregation in Cu, Zn superoxide dismutase: a molecular dynamics simulation study.J Biomol Struct Dyn. 2018 Aug;36(10):2605-2617. doi: 10.1080/07391102.2017.1364670. Epub 2017 Aug 20. J Biomol Struct Dyn. 2018. PMID: 28782426
-
Unveiling the unfolding pathway of FALS associated G37R SOD1 mutant: a computational study.Mol Biosyst. 2010 Jun;6(6):1032-9. doi: 10.1039/b918662j. Epub 2010 Feb 23. Mol Biosyst. 2010. PMID: 20485746
-
TFE-induced local unfolding and fibrillation of SOD1: bridging the experiment and simulation studies.Biochem J. 2018 May 18;475(10):1701-1719. doi: 10.1042/BCJ20180085. Biochem J. 2018. PMID: 29686043
-
Mutant SOD1 instability: implications for toxicity in amyotrophic lateral sclerosis.Neurodegener Dis. 2005;2(3-4):115-27. doi: 10.1159/000089616. Neurodegener Dis. 2005. PMID: 16909016 Review.
-
SOD1 aggregation and ALS: role of metallation states and disulfide status.Curr Top Med Chem. 2012;12(22):2560-72. doi: 10.2174/1568026611212220010. Curr Top Med Chem. 2012. PMID: 23339308 Review.
Cited by
-
A structural modeling approach for the understanding of initiation and elongation of ALS-linked superoxide dismutase fibrils.J Mol Model. 2013 Sep;19(9):3695-704. doi: 10.1007/s00894-013-1896-7. Epub 2013 Jun 19. J Mol Model. 2013. PMID: 23780345
-
Conformational dynamics of superoxide dismutase (SOD1) in osmolytes: a molecular dynamics simulation study.RSC Adv. 2020 Jul 30;10(46):27598-27614. doi: 10.1039/d0ra02151b. eCollection 2020 Jul 21. RSC Adv. 2020. PMID: 35516947 Free PMC article.
-
Inhibition of IAPP aggregation by insulin depends on the insulin oligomeric state regulated by zinc ion concentration.Sci Rep. 2015 Feb 4;5:8240. doi: 10.1038/srep08240. Sci Rep. 2015. PMID: 25649462 Free PMC article.
-
Nanosecond pulsed electric signals can affect electrostatic environment of proteins below the threshold of conformational effects: The case study of SOD1 with a molecular simulation study.PLoS One. 2019 Aug 27;14(8):e0221685. doi: 10.1371/journal.pone.0221685. eCollection 2019. PLoS One. 2019. PMID: 31454403 Free PMC article.
-
SOD1-positive aggregate accumulation in the CNS predicts slower disease progression and increased longevity in a mutant SOD1 mouse model of ALS.Sci Rep. 2019 Apr 30;9(1):6724. doi: 10.1038/s41598-019-43164-z. Sci Rep. 2019. PMID: 31040321 Free PMC article.
References
-
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. - PubMed
-
- Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu Rev Neurosci. 2004;27:723–49. - PubMed
-
- Oztug Durer ZA, Cohlberg JA, Dinh P, Padua S, Ehrenclou K, Downes S, Tan JK, Nakano Y, Bowman CJ, Hoskins JL, Kwon C, Mason AZ, Rodriguez JA, Doucette PA, Shaw BF, Selverstone Valentine J. Loss of metal ions, disulfide reduction and mutations related to familial ALS promote formation of amyloid-like aggregates from superoxide dismutase. PLoS One. 2009;4:e5004. - PMC - PubMed
-
- Jahn TR, Makin OS, Morris KL, Marshall KE, Tian P, Sikorski P, Serpell LC. The common architecture of cross-beta amyloid. J Mol Biol. 2010;395:717–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

