On marathons and Sprints: an integrated quantitative proteomics and transcriptomics analysis of differences between slow and fast muscle fibers
- PMID: 22210690
- PMCID: PMC3433927
- DOI: 10.1074/mcp.M111.010801
On marathons and Sprints: an integrated quantitative proteomics and transcriptomics analysis of differences between slow and fast muscle fibers
Abstract
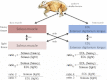
Skeletal muscle tissue contains slow as well as fast twitch muscle fibers that possess different metabolic and contractile properties. Although the distribution of individual proteins in fast and slow fibers has been investigated extensively, a comprehensive proteomic analysis, which is key for any systems biology approach to muscle tissues, is missing. Here, we compared the global protein levels and gene expression profiles of the predominantly slow soleus and fast extensor digitorum longus muscles using the principle of in vivo stable isotope labeling with amino acids based on a fully lysine-6 labeled SILAC-mouse. We identified 551 proteins with significant quantitative differences between slow soleus and fast extensor digitorum longus fibers out of >2000 quantified proteins, which greatly extends the repertoire of proteins differentially regulated between both muscle types. Most of the differentially regulated proteins mediate cellular contraction, ion homeostasis, glycolysis, and oxidation, which reflect the major functional differences between both muscle types. Comparison of proteomics and transcriptomics data uncovered the existence of fiber-type specific posttranscriptional regulatory mechanisms resulting in differential accumulation of Myosin-8 and α-protein kinase 3 proteins and mRNAs among others. Phosphoproteome analysis of soleus and extensor digitorum longus muscles identified 2573 phosphosites on 973 proteins including 1040 novel phosphosites. The in vivo stable isotope labeling with amino acids-mouse approach used in our study provides a comprehensive view into the protein networks that direct fiber-type specific functions and allows a detailed dissection of the molecular composition of slow and fast muscle tissues with unprecedented resolution.
Figures




References
-
- Schiaffino S., Reggiani C. (1996) Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol. Rev. 76, 371–423 - PubMed
-
- Booth F. W., Thomason D. B. (1991) Molecular and cellular adaptation of muscle in response to exercise: perspectives of various models. Physiol. Rev. 71, 541–585 - PubMed
-
- Pette D., Staron R. S. (2000) Myosin isoforms, muscle fiber types, and transitions. Microsc. Res. Tech. 50, 500–509 - PubMed
-
- Dhoot G. K., Perry S. V. (1979) Distribution of polymorphic forms of troponin components and tropomyosin in skeletal muscle. Nature 278, 714–718 - PubMed
-
- MacLennan D. H., Toyofuku T., Lytton J. (1992) Structure-function relationships in sarcoplasmic or endoplasmic reticulum type Ca2+ pumps. Ann. N.Y. Acad. Sci. 671, 1–10 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

