iTRAQ labeling is superior to mTRAQ for quantitative global proteomics and phosphoproteomics
- PMID: 22210691
- PMCID: PMC3433912
- DOI: 10.1074/mcp.M111.014423
iTRAQ labeling is superior to mTRAQ for quantitative global proteomics and phosphoproteomics
Abstract
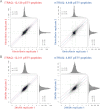
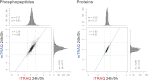
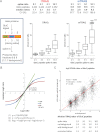
Labeling of primary amines on peptides with reagents containing stable isotopes is a commonly used technique in quantitative mass spectrometry. Isobaric labeling techniques such as iTRAQ™ or TMT™ allow for relative quantification of peptides based on ratios of reporter ions in the low m/z region of spectra produced by precursor ion fragmentation. In contrast, nonisobaric labeling with mTRAQ™ yields precursors with different masses that can be directly quantified in MS1 spectra. In this study, we compare iTRAQ- and mTRAQ-based quantification of peptides and phosphopeptides derived from EGF-stimulated HeLa cells. Both labels have identical chemical structures, therefore precursor ion- and fragment ion-based quantification can be directly compared. Our results indicate that iTRAQ labeling has an additive effect on precursor intensities, whereas mTRAQ labeling leads to more redundant MS2 scanning events caused by triggering on the same peptide with different mTRAQ labels. We found that iTRAQ labeling quantified nearly threefold more phosphopeptides (12,129 versus 4,448) and nearly twofold more proteins (2,699 versus 1,597) than mTRAQ labeling. Although most key proteins in the EGFR signaling network were quantified with both techniques, iTRAQ labeling allowed quantification of twice as many kinases. Accuracy of reporter ion quantification by iTRAQ is adversely affected by peptides that are cofragmented in the same precursor isolation window, dampening observed ratios toward unity. However, because of tighter overall iTRAQ ratio distributions, the percentage of statistically significantly regulated phosphopeptides and proteins detected by iTRAQ and mTRAQ was similar. We observed a linear correlation of logarithmic iTRAQ to mTRAQ ratios over two orders of magnitude, indicating a possibility to correct iTRAQ ratios by an average compression factor. Spike-in experiments using peptides of defined ratios in a background of nonregulated peptides show that iTRAQ quantification is less accurate but not as variable as mTRAQ quantification.
Figures



References
-
- Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 - PubMed
-
- Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., Hamon C. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 - PubMed
-
- Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 - PubMed
-
- Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

