Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein
- PMID: 22210774
- PMCID: PMC3293566
- DOI: 10.1074/jbc.M111.295121
Structure-guided engineering enhances a phytochrome-based infrared fluorescent protein
Abstract
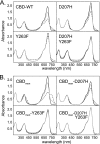
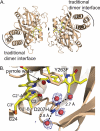
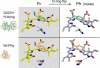
Phytochrome is a multidomain dimeric red light photoreceptor that utilizes a chromophore-binding domain (CBD), a PHY domain, and an output module to induce cellular changes in response to light. A promising biotechnology tool emerged when a structure-based substitution at Asp-207 was shown to be an infrared fluorophore that uses a biologically available tetrapyrrole chromophore. We report multiple crystal structures of this D207H variant of the Deinococcus radiodurans CBD, in which His-207 is observed to form a hydrogen bond with either the tetrapyrrole A-ring oxygen or the Tyr-263 hydroxyl. Based on the implications of this duality for fluorescence properties, Y263F was introduced and shown to have stronger fluorescence than the original D207H template. Our structures are consistent with the model that the Y263F change prevents a red light-induced far-red light absorbing phytochrome chromophore configuration. With the goal of decreasing size and thereby facilitating use as a fluorescent tag in vivo, we also engineered a monomeric form of the CBD. Unexpectedly, photoconversion was observed in the monomer despite the lack of a PHY domain. This observation underscores an interplay between dimerization and the photochemical properties of phytochrome and suggests that the monomeric CBD could be used for further studies of the photocycle. The D207H substitution on its own in the monomer did not result in fluorescence, whereas Y263F did. Combined, the D207H and Y263F substitutions in the monomeric CBD lead to the brightest of our variants, designated Wisconsin infrared phytofluor (Wi-Phy).
Figures



Similar articles
-
On the (un)coupling of the chromophore, tongue interactions, and overall conformation in a bacterial phytochrome.J Biol Chem. 2018 May 25;293(21):8161-8172. doi: 10.1074/jbc.RA118.001794. Epub 2018 Apr 5. J Biol Chem. 2018. PMID: 29622676 Free PMC article.
-
Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes.J Biol Chem. 2008 May 2;283(18):12212-26. doi: 10.1074/jbc.M709355200. Epub 2008 Jan 10. J Biol Chem. 2008. PMID: 18192276 Free PMC article.
-
Crystallographic and electron microscopic analyses of a bacterial phytochrome reveal local and global rearrangements during photoconversion.J Biol Chem. 2014 Aug 29;289(35):24573-87. doi: 10.1074/jbc.M114.571661. Epub 2014 Jul 8. J Biol Chem. 2014. PMID: 25006244 Free PMC article.
-
Spectroscopy and a high-resolution crystal structure of Tyr263 mutants of cyanobacterial phytochrome Cph1.J Mol Biol. 2011 Oct 14;413(1):115-27. doi: 10.1016/j.jmb.2011.08.023. Epub 2011 Aug 23. J Mol Biol. 2011. PMID: 21888915
-
Bacterial Phytochromes, Cyanobacteriochromes and Allophycocyanins as a Source of Near-Infrared Fluorescent Probes.Int J Mol Sci. 2017 Aug 3;18(8):1691. doi: 10.3390/ijms18081691. Int J Mol Sci. 2017. PMID: 28771184 Free PMC article. Review.
Cited by
-
Modulation of Structural Heterogeneity Controls Phytochrome Photoswitching.Biophys J. 2020 Jan 21;118(2):415-421. doi: 10.1016/j.bpj.2019.11.025. Epub 2019 Nov 26. Biophys J. 2020. PMID: 31839260 Free PMC article.
-
Advances in whole-embryo imaging: a quantitative transition is underway.Nat Rev Mol Cell Biol. 2014 May;15(5):327-39. doi: 10.1038/nrm3786. Epub 2014 Apr 16. Nat Rev Mol Cell Biol. 2014. PMID: 24739741 Review.
-
Structural insights into photoactivation and signalling in plant phytochromes.Nat Plants. 2020 May;6(5):581-588. doi: 10.1038/s41477-020-0638-y. Epub 2020 May 4. Nat Plants. 2020. PMID: 32366982
-
A naturally monomeric infrared fluorescent protein for protein labeling in vivo.Nat Methods. 2015 Aug;12(8):763-5. doi: 10.1038/nmeth.3447. Epub 2015 Jun 22. Nat Methods. 2015. PMID: 26098020 Free PMC article.
-
Comparative Analysis of Bacteriophytochrome Agp2 and Its Engineered Photoactivatable NIR Fluorescent Proteins PAiRFP1 and PAiRFP2.Biomolecules. 2020 Sep 7;10(9):1286. doi: 10.3390/biom10091286. Biomolecules. 2020. PMID: 32906690 Free PMC article.
References
-
- Auldridge M. E., Forest K. T. (2011) Bacterial phytochromes: more than meets the light. Crit. Rev. Biochem. Mol. Biol. 46, 67–88 - PubMed
-
- Schumann C., Gross R., Michael N., Lamparter T., Diller R. (2007) Sub-picosecond mid-infrared spectroscopy of phytochrome Agp1 from Agrobacterium tumefaciens. ChemPhysChem 8, 1657–1663 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

