Interplay of a ligand sensor and an enzyme in controlling expression of the Saccharomyces cerevisiae GAL genes
- PMID: 22210830
- PMCID: PMC3294446
- DOI: 10.1128/EC.05294-11
Interplay of a ligand sensor and an enzyme in controlling expression of the Saccharomyces cerevisiae GAL genes
Abstract
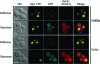
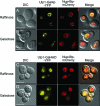
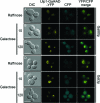
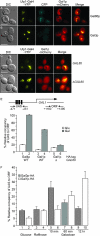
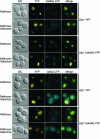
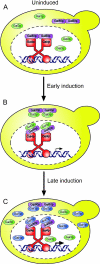
The regulation of the Saccharomyces cerevisiae GAL genes in response to galactose as a source of carbon has served as a paradigm for eukaryotic transcriptional control over the last 50 years. Three proteins--a transcriptional activator (Gal4p), an inhibitor (Gal80p), and a ligand sensor (Gal3p)--control the switch between inert and active gene expression. The molecular mechanism by which the recognition of galactose within the cell is converted into a transcriptional response has been the subject of considerable debate. In this study, using a novel and powerful method of localizing active transcription factors within the nuclei of cells, we show that a short-lived complex between Gal4p, Gal80p, and Gal3p occurs soon after the addition of galactose to cells to activate GAL gene expression. Gal3p is subsequently replaced in this complex by Gal1p, and a Gal4p-Gal80p-Gal1p complex is responsible for the continued expression of the GAL genes. The transient role of the ligand sensor indicates that current models for the induction and continued expression of the yeast GAL genes need to be reevaluated.
Figures
References
-
- Barberis A, et al. 1995. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell 81:359–368 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

