In vitro experimental system for analysis of transcription-translation coupling
- PMID: 22210860
- PMCID: PMC3315309
- DOI: 10.1093/nar/gkr1262
In vitro experimental system for analysis of transcription-translation coupling
Abstract
Transcription and translation are coupled in bacteria, meaning that translation takes place co-transcriptionally. During transcription-translation, both machineries mutually affect each others' functions, which is important for regulation of gene expression. Analysis of interactions between RNA polymerase (RNAP) and the ribosome, however, are limited due to the lack of an in vitro experimental system. Here, we report the development of an in vitro transcription coupled to translation system assembled from purified components. The system allows controlled stepwise transcription and simultaneous stepwise translation of the nascent RNA, and permits investigation of the interactions of RNAP with the ribosome, as well as the effects of translation on transcription and transcription on translation. As an example of usage of this experimental system, we uncover complex effects of transcription-translation coupling on pausing of transcription.
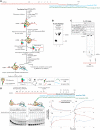
Figures



References
-
- Zenkin N, Naryshkina T, Kuznedelov K, Severinov K. The mechanism of DNA replication primer synthesis by RNA polymerase. Nature. 2006;439:617–620. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

