Lactoferrin inhibits Porphyromonas gingivalis proteinases and has sustained biofilm inhibitory activity
- PMID: 22214780
- PMCID: PMC3294922
- DOI: 10.1128/AAC.05100-11
Lactoferrin inhibits Porphyromonas gingivalis proteinases and has sustained biofilm inhibitory activity
Abstract
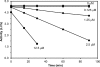
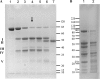
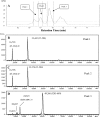
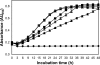
Porphyromonas gingivalis is a bacterial pathogen associated with chronic periodontitis that results in destruction of the tooth's supporting tissues. The major virulence determinants of P. gingivalis are its cell surface Arg- and Lys-specific cysteine proteinases, RgpA/B and Kgp. Lactoferrin (LF), an 80-kDa iron-binding glycoprotein found in saliva and gingival crevicular fluid, is believed to play an important role in innate immunity. In this study, bovine milk LF displayed proteinase inhibitory activity against P. gingivalis whole cells, significantly inhibiting both Arg- and Lys-specific proteolytic activities. LF inhibited the Arg-specific activity of purified RgpB, which lacks adhesin domains, and also inhibited the same activity of the RgpA/Kgp proteinase-adhesin complexes in a time-dependent manner, with a first-order inactivation rate constant (k(inact)) of 0.023 min(-1) and an inhibitor affinity constant (K(I)) of 5.02 μM. LF inhibited P. gingivalis biofilm formation by >80% at concentrations above 0.625 μM. LF was relatively resistant to hydrolysis by P. gingivalis cells but was cleaved into two major polypeptides (53 and 33 kDa) at R(284) to S(285), as determined by in-source decay mass spectrometry; however, these polypeptides remained associated with each other and retained inhibitory activity. The biofilm inhibitory activity of LF against P. gingivalis was not attributed to direct antibacterial activity, as LF displayed little growth inhibitory activity against planktonic cells. As the known RgpA/B and Kgp inhibitor N-α-p-tosyl-l-lysine chloromethylketone also inhibited P. gingivalis biofilm formation, the antibiofilm effect of LF may at least in part be attributable to its antiproteinase activity.
Figures





References
-
- Albandar JM, Brunelle JA, Kingman A. 1999. Destructive periodontal disease in adults 30 years of age and older in the United States, 1988-1994. J. Periodontol. 70:13–29 - PubMed
-
- Armfield J, Roberts-Thomson K, Spencer A. 2000. Australia's health 2000, vol 7. Catalogue no. AUS 19. Australian Institute of Health and Welfare, Canberra, Australia
-
- Bhogal PS, Slakeski N, Reynolds EC. 1997. A cell-associated protein complex of Porphyromonas gingivalis W50 composed of Arg- and Lys-specific cysteine proteinases and adhesins. Microbiology 143:2485–2495 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

