A novel feedback loop regulates the response to endoplasmic reticulum stress via the cooperation of cytoplasmic splicing and mRNA translation
- PMID: 22215619
- PMCID: PMC3295193
- DOI: 10.1128/MCB.06665-11
A novel feedback loop regulates the response to endoplasmic reticulum stress via the cooperation of cytoplasmic splicing and mRNA translation
Abstract
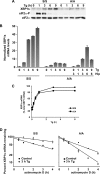
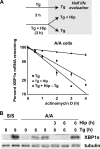
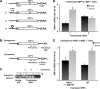
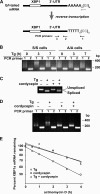
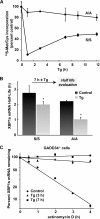
The accumulation of unfolded proteins in the endoplasmic reticulum (ER) triggers transcriptional and translational reprogramming. This unfolded protein response (UPR) protects cells during transient stress and can lead to apoptosis during prolonged stress. Two key mediators of the UPR are PKR-like ER kinase (PERK), which phosphorylates the α subunit of eukaryotic translation initiation factor 2 (eIF2α), resulting in decreased protein synthesis, and the α subunit of inositol-requiring enzyme 1 (IRE1α), which initiates cytoplasmic splicing of the mRNA encoding the transcription factor X-box binding protein 1 (XBP1). XBP1 induces transcription of genes involved in protein quality control. This report describes cross talk between these two pathways: phosphorylation of eIF2α was required for maximal induction of spliced XBP1 (XBP1s) protein levels via a mechanism that involved stabilization of XBP1s mRNA. By using mouse embryo fibroblasts deficient in UPR signaling pathways, we demonstrate that stress-induced stabilization of XBP1s mRNA requires cytoplasmic splicing of the mRNA and inhibition of its translation. Because the XBP1s protein promotes transcription of its own gene, the UPR-induced mRNA stabilization is part of a positive feedback loop that induces XBP1s protein accumulation and transcription of target genes during stress. We propose a model in which eIF2α phosphorylation-mediated control of mRNA turnover is a molecular switch that regulates the stress response transcription program and the ER's capacity for protein folding during stress.
Figures

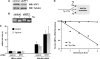
References
-
- Adachi Y, et al. 2008. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct. Funct 33:75–89 - PubMed
-
- Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. 2000. Dynamic interaction of BiP and ER stress transducers in the unfolded- protein response. Nat. Cell Biol. 2:326–332 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
