Genome comparison of human and non-human malaria parasites reveals species subset-specific genes potentially linked to human disease
- PMID: 22215999
- PMCID: PMC3245289
- DOI: 10.1371/journal.pcbi.1002320
Genome comparison of human and non-human malaria parasites reveals species subset-specific genes potentially linked to human disease
Abstract
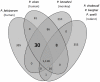
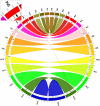
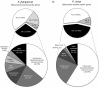
Genes underlying important phenotypic differences between Plasmodium species, the causative agents of malaria, are frequently found in only a subset of species and cluster at dynamically evolving subtelomeric regions of chromosomes. We hypothesized that chromosome-internal regions of Plasmodium genomes harbour additional species subset-specific genes that underlie differences in human pathogenicity, human-to-human transmissibility, and human virulence. We combined sequence similarity searches with synteny block analyses to identify species subset-specific genes in chromosome-internal regions of six published Plasmodium genomes, including Plasmodium falciparum, Plasmodium vivax, Plasmodium knowlesi, Plasmodium yoelii, Plasmodium berghei, and Plasmodium chabaudi. To improve comparative analysis, we first revised incorrectly annotated gene models using homology-based gene finders and examined putative subset-specific genes within syntenic contexts. Confirmed subset-specific genes were then analyzed for their role in biological pathways and examined for molecular functions using publicly available databases. We identified 16 genes that are well conserved in the three primate parasites but not found in rodent parasites, including three key enzymes of the thiamine (vitamin B1) biosynthesis pathway. Thirteen genes were found to be present in both human parasites but absent in the monkey parasite P. knowlesi, including genes specifically upregulated in sporozoites or gametocytes that could be linked to parasite transmission success between humans. Furthermore, we propose 15 chromosome-internal P. falciparum-specific genes as new candidate genes underlying increased human virulence and detected a currently uncharacterized cluster of P. vivax-specific genes on chromosome 6 likely involved in erythrocyte invasion. In conclusion, Plasmodium species harbour many chromosome-internal differences in the form of protein-coding genes, some of which are potentially linked to human disease and thus promising leads for future laboratory research.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- WHO. World Malaria Report 2008. World Health Organization; 2008.
-
- Cox FEG. Studies on the host parasite relationships of Haemosporidia. 1964. [Ph. D. Thesis]: University of London.
-
- Killick-Kendrick R, Peters W. Rodent Malaria. London: Academic Press Inc. Ltd; 1978. 62
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

