Restricting microbial exposure in early life negates the immune benefits associated with gut colonization in environments of high microbial diversity
- PMID: 22216092
- PMCID: PMC3245219
- DOI: 10.1371/journal.pone.0028279
Restricting microbial exposure in early life negates the immune benefits associated with gut colonization in environments of high microbial diversity
Abstract
Background: Acquisition of the intestinal microbiota in early life corresponds with the development of the mucosal immune system. Recent work on caesarean-delivered infants revealed that early microbial composition is influenced by birthing method and environment. Furthermore, we have confirmed that early-life environment strongly influences both the adult gut microbiota and development of the gut immune system. Here, we address the impact of limiting microbial exposure after initial colonization on the development of adult gut immunity.
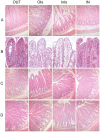
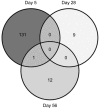
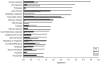
Methodology/principal findings: Piglets were born in indoor or outdoor rearing units, allowing natural colonization in the immediate period after birth, prior to transfer to high-health status isolators. Strikingly, gut closure and morphological development were strongly affected by isolator-rearing, independent of indoor or outdoor origins of piglets. Isolator-reared animals showed extensive vacuolation and disorganization of the gut epithelium, inferring that normal gut closure requires maturation factors present in maternal milk. Although morphological maturation and gut closure were delayed in isolator-reared animals, these hard-wired events occurred later in development. Type I IFN, IL-22, IL-23 and Th17 pathways were increased in indoor-isolator compared to outdoor-isolator animals during early life, indicating greater immune activation in pigs originating from indoor environments reflecting differences in the early microbiota. This difference was less apparent later in development due to enhanced immune activation and convergence of the microbiota in all isolator-reared animals. This correlated with elevation of Type I IFN pathways in both groups, although T cell pathways were still more affected in indoor-reared animals.
Conclusions/significance: Environmental factors, in particular microbial exposure, influence expression of a large number of immune-related genes. However, the homeostatic effects of microbial colonization in outdoor environments require sustained microbial exposure throughout development. Gut development in high-hygiene environments negatively impacts on normal succession of the gut microbiota and promotes innate immune activation which may impair immune homeostasis.
© 2011 Mulder et al.
Conflict of interest statement
Figures
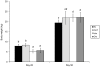

References
-
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–848. - PubMed
-
- Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9:279–290. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

