Replication, gene expression and particle production by a consensus Merkel Cell Polyomavirus (MCPyV) genome
- PMID: 22216177
- PMCID: PMC3246459
- DOI: 10.1371/journal.pone.0029112
Replication, gene expression and particle production by a consensus Merkel Cell Polyomavirus (MCPyV) genome
Abstract
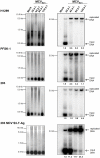
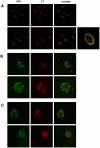
Merkel Cell Polyomavirus (MCPyV) genomes are clonally integrated in tumor tissues of approximately 85% of all Merkel cell carcinoma (MCC) cases, a highly aggressive tumor of the skin which predominantly afflicts elderly and immunosuppressed patients. All integrated viral genomes recovered from MCC tissue or MCC cell lines harbor signature mutations in the early gene transcript encoding for the large T-Antigen (LT-Ag). These mutations selectively abrogate the ability of LT-Ag to support viral replication while still maintaining its Rb-binding activity, suggesting a continuous requirement for LT-Ag mediated cell cycle deregulation during MCC pathogenesis. To gain a better understanding of MCPyV biology, in vitro MCPyV replication systems are required. We have generated a synthetic MCPyV genomic clone (MCVSyn) based on the consensus sequence of MCC-derived sequences deposited in the NCBI database. Here, we demonstrate that transfection of recircularized MCVSyn DNA into some human cell lines recapitulates efficient replication of the viral genome, early and late gene expression together with virus particle formation. However, serial transmission of infectious virus was not observed. This in vitro culturing system allows the study of viral replication and will facilitate the molecular dissection of important aspects of the MCPyV lifecycle.
© 2011 Neumann et al.
Conflict of interest statement
Figures



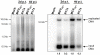
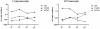


References
-
- Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1:1253–1257. - PubMed
-
- Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet. 1971;1:1257–1260. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

