Heterologous expression of membrane proteins: choosing the appropriate host
- PMID: 22216205
- PMCID: PMC3244453
- DOI: 10.1371/journal.pone.0029191
Heterologous expression of membrane proteins: choosing the appropriate host
Abstract
Background: Membrane proteins are the targets of 50% of drugs, although they only represent 1% of total cellular proteins. The first major bottleneck on the route to their functional and structural characterisation is their overexpression; and simply choosing the right system can involve many months of trial and error. This work is intended as a guide to where to start when faced with heterologous expression of a membrane protein.
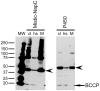
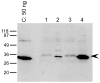
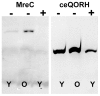
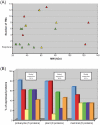
Methodology/principal findings: The expression of 20 membrane proteins, both peripheral and integral, in three prokaryotic (E. coli, L. lactis, R. sphaeroides) and three eukaryotic (A. thaliana, N. benthamiana, Sf9 insect cells) hosts was tested. The proteins tested were of various origins (bacteria, plants and mammals), functions (transporters, receptors, enzymes) and topologies (between 0 and 13 transmembrane segments). The Gateway system was used to clone all 20 genes into appropriate vectors for the hosts to be tested. Culture conditions were optimised for each host, and specific strategies were tested, such as the use of Mistic fusions in E. coli. 17 of the 20 proteins were produced at adequate yields for functional and, in some cases, structural studies. We have formulated general recommendations to assist with choosing an appropriate system based on our observations of protein behaviour in the different hosts.
Conclusions/significance: Most of the methods presented here can be quite easily implemented in other laboratories. The results highlight certain factors that should be considered when selecting an expression host. The decision aide provided should help both newcomers and old-hands to select the best system for their favourite membrane protein.
© 2011 Bernaudat et al.
Conflict of interest statement
Figures


References
-
- Lacapère J-J, Pebay-Peyroula E, Neumann J-M, Etchebest C. Determining membrane protein structures: still a challenge! Trends Biochem Sci. 2007;32:259–270. doi: 10.1016/j.tibs.2007.04.001. - DOI - PubMed
-
- von Heijne G. The membrane protein universe: what's out there and why bother? J Intern Med. 2007;261:543–557. doi: 10.1111/j.1365-2796.2007.01792.x. - DOI - PubMed
-
- McLuskey K, Roszak AW, Zhu Y, Isaacs NW. Crystal structures of all-alpha type membrane proteins. Eur Biophys J. 2010;39:723–755. doi: 10.1007/s00249-009-0546-6. - DOI - PubMed
-
- Wagner S, Bader ML, Drew D, de Gier J-W. Rationalizing membrane protein overexpression. Trends Biotechnol. 2006;24:364–371. doi: 10.1016/j.tibtech.2006.06.008. - DOI - PubMed
-
- White SH. Biophysical dissection of membrane proteins. Nature. 2009;459:344–346. doi: 10.1038/nature08142. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

