Preservation of ranking order in the expression of human Housekeeping genes
- PMID: 22216246
- PMCID: PMC3245260
- DOI: 10.1371/journal.pone.0029314
Preservation of ranking order in the expression of human Housekeeping genes
Abstract
Housekeeping (HK) genes fulfill the basic needs for a cell to survive and function properly. Their ubiquitous expression, originally thought to be constant, can vary from tissue to tissue, but this variation remains largely uncharacterized and it could not be explained by previously identified properties of HK genes such as short gene length and high GC content. By analyzing microarray expression data for human genes, we uncovered a previously unnoted characteristic of HK gene expression, namely that the ranking order of their expression levels tends to be preserved from one tissue to another. Further analysis by tensor product decomposition and pathway stratification identified three main factors of the observed ranking preservation, namely that, compared to those of non-HK (NHK) genes, the expression levels of HK genes show a greater degree of dispersion (less overlap), stableness (a smaller variation in expression between tissues), and correlation of expression. Our results shed light on regulatory mechanisms of HK gene expression that are probably different for different HK genes or pathways, but are consistent and coordinated in different tissues.
© 2011 Shaw et al.
Conflict of interest statement
Figures
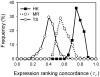
 ). Each distribution is a compilation of
). Each distribution is a compilation of  630 Kendall's tau (
630 Kendall's tau ( ; Equation (3
)), where each
; Equation (3
)), where each  was for a pair of tissues sampled from a 36-tissue pool and the frequency of
was for a pair of tissues sampled from a 36-tissue pool and the frequency of  at a particular value was computed on a histogram using a bin width of 0.1. The average value (
at a particular value was computed on a histogram using a bin width of 0.1. The average value ( ) of the
) of the  distribution for the HK, MR, and TS gene sets was 0.77, 0.59, and 0.41, respectively, and the standard errors were all small, mostly less than 0.01.
distribution for the HK, MR, and TS gene sets was 0.77, 0.59, and 0.41, respectively, and the standard errors were all small, mostly less than 0.01.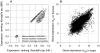
 computed for the two gene groups (Ng = 388 and Ng = 11,953) for a particular pair of tissues. (B) The expression of all the genes in any two human tissues (heart and thymus are shown as an example) always has an elliptical shape, resulting in a substantially preserved ranking order with a non-zero
computed for the two gene groups (Ng = 388 and Ng = 11,953) for a particular pair of tissues. (B) The expression of all the genes in any two human tissues (heart and thymus are shown as an example) always has an elliptical shape, resulting in a substantially preserved ranking order with a non-zero  (>0.4) even for randomly grouped genes (triangles in (A)). In contrast, randomly assigned rankings would yield a
(>0.4) even for randomly grouped genes (triangles in (A)). In contrast, randomly assigned rankings would yield a  value very close to zero (squares in (A)). Gene expression levels were log2 transformed and were denoted by xgt.
value very close to zero (squares in (A)). Gene expression levels were log2 transformed and were denoted by xgt.

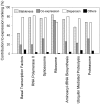
 ) computed for the HK genes found in each of the seven HK-enriched KEGG pathways. The black bars are the contributions of other unknown factors.
) computed for the HK genes found in each of the seven HK-enriched KEGG pathways. The black bars are the contributions of other unknown factors.


 ) computed for HK genes coding for proteins involved in different subcomplexes of the proteasome. The
) computed for HK genes coding for proteins involved in different subcomplexes of the proteasome. The  value is shown in parenthesis at the bottom of the box. * indicates that the value is significantly different from
value is shown in parenthesis at the bottom of the box. * indicates that the value is significantly different from  computed from random sampling using the same number of genes.
computed from random sampling using the same number of genes.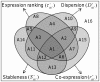
 ) and the three contributing factors of co-expression (
) and the three contributing factors of co-expression ( , Equation (5
)), stableness (
, Equation (5
)), stableness ( , Equation (7
)), and dispersion (
, Equation (7
)), and dispersion ( , Equation (8
)). The rectangular box represents the universal set, its 16 components can be deduced from tensor computations (Equation (9
)), e.g., A8 =
, Equation (8
)). The rectangular box represents the universal set, its 16 components can be deduced from tensor computations (Equation (9
)), e.g., A8 =  , A10 =
, A10 =  , and A16 =
, and A16 =  . The equations for computing these components are described in the Methods (Equation (11
–
13
)). Note that each of the four elements (
. The equations for computing these components are described in the Methods (Equation (11
–
13
)). Note that each of the four elements ( ,
, ,
, , and
, and  ) is a composite of eight components.
) is a composite of eight components.Similar articles
-
Partitioning the human transcriptome using HKera, a novel classifier of housekeeping and tissue-specific genes.PLoS One. 2013 Dec 20;8(12):e83040. doi: 10.1371/journal.pone.0083040. eCollection 2013. PLoS One. 2013. PMID: 24376628 Free PMC article.
-
HRT Atlas v1.0 database: redefining human and mouse housekeeping genes and candidate reference transcripts by mining massive RNA-seq datasets.Nucleic Acids Res. 2021 Jan 8;49(D1):D947-D955. doi: 10.1093/nar/gkaa609. Nucleic Acids Res. 2021. PMID: 32663312 Free PMC article.
-
Un-biased housekeeping gene panel selection for high-validity gene expression analysis.Sci Rep. 2022 Jul 19;12(1):12324. doi: 10.1038/s41598-022-15989-8. Sci Rep. 2022. PMID: 35853974 Free PMC article.
-
DNA entropy reveals a significant difference in complexity between housekeeping and tissue specific gene promoters.Comput Biol Chem. 2015 Oct;58:19-24. doi: 10.1016/j.compbiolchem.2015.05.001. Epub 2015 May 8. Comput Biol Chem. 2015. PMID: 25988219
-
Validation of Housekeeping Genes for Gene Expression Profiling in Fish: A Necessity.Crit Rev Eukaryot Gene Expr. 2019;29(6):565-579. doi: 10.1615/CritRevEukaryotGeneExpr.2019028964. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422011 Review.
Cited by
-
Brain areas affected by intranasal oxytocin show higher oxytocin receptor expression.Eur J Neurosci. 2021 Oct;54(7):6374-6381. doi: 10.1111/ejn.15447. Epub 2021 Sep 16. Eur J Neurosci. 2021. PMID: 34498316 Free PMC article.
-
Familial or Sporadic Idiopathic Scoliosis - classification based on artificial neural network and GAPDH and ACTB transcription profile.Biomed Eng Online. 2013 Jan 4;12:1. doi: 10.1186/1475-925X-12-1. Biomed Eng Online. 2013. PMID: 23289769 Free PMC article.
-
Transcriptional and cell type profiles of cortical brain regions showing ultradian cortisol rhythm dependent responses to emotional face stimulation.Neurobiol Stress. 2023 Jan 4;22:100514. doi: 10.1016/j.ynstr.2023.100514. eCollection 2023 Jan. Neurobiol Stress. 2023. PMID: 36660181 Free PMC article.
-
Canonical genetic signatures of the adult human brain.Nat Neurosci. 2015 Dec;18(12):1832-44. doi: 10.1038/nn.4171. Epub 2015 Nov 16. Nat Neurosci. 2015. PMID: 26571460 Free PMC article.
-
Partitioning the human transcriptome using HKera, a novel classifier of housekeeping and tissue-specific genes.PLoS One. 2013 Dec 20;8(12):e83040. doi: 10.1371/journal.pone.0083040. eCollection 2013. PLoS One. 2013. PMID: 24376628 Free PMC article.
References
-
- Watson JD, Hopkins NH, Roberts JW, Steitz JA, Weiner AM. The functioning of higher eucaryotic genes. Molecular biology of the gene. 1 ed. Menlo Park, , California: Benjamin/Cummings; 1965. 704
-
- Yamada H, Chen D, Monstein HJ, Hakanson R. Effects of fasting on the expression of gastrin, cholecystokinin, and somatostatin genes and of various housekeeping genes in the pancreas and upper digestive tract of rats. Biochem Biophys Res Commun. 1997;231:835–838. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

