Multiple polymorphisms affect expression and function of the neuropeptide S receptor (NPSR1)
- PMID: 22216302
- PMCID: PMC3244468
- DOI: 10.1371/journal.pone.0029523
Multiple polymorphisms affect expression and function of the neuropeptide S receptor (NPSR1)
Abstract
Background: neuropeptide S (NPS) and its receptor NPSR1 act along the hypothalamic-pituitary-adrenal axis to modulate anxiety, fear responses, nociception and inflammation. The importance of the NPS-NPSR1 signaling pathway is highlighted by the observation that, in humans, NPSR1 polymorphism associates with asthma, inflammatory bowel disease, rheumatoid arthritis, panic disorders, and intermediate phenotypes of functional gastrointestinal disorders. Because of the genetic complexity at the NPSR1 locus, however, true causative variations remain to be identified, together with their specific effects on receptor expression or function. To gain insight into the mechanisms leading to NPSR1 disease-predisposing effects, we performed a thorough functional characterization of all NPSR1 promoter and coding SNPs commonly occurring in Caucasians (minor allele frequency >0.02).
Principal findings: we identified one promoter SNP (rs2530547 [-103]) that significantly affects luciferase expression in gene reporter assays and NPSR1 mRNA levels in human leukocytes. We also detected quantitative differences in NPS-induced genome-wide transcriptional profiles and CRE-dependent luciferase activities associated with three NPSR1 non-synonymous SNPs (rs324981 [Ile107Asn], rs34705969 [Cys197Phe], rs727162 [Arg241Ser]), with a coding variant exhibiting a loss-of-function phenotype (197Phe). Potential mechanistic explanations were sought with molecular modelling and bioinformatics, and a pilot study of 2230 IBD cases and controls provided initial support to the hypothesis that different cis-combinations of these functional SNPs variably affect disease risk.
Significance: these findings represent a first step to decipher NPSR1 locus complexity and its impact on several human conditions NPS antagonists have been recently described, and our results are of potential pharmacogenetic relevance.
© 2011 Anedda et al.
Conflict of interest statement
Figures

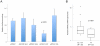


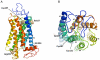
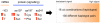
References
-
- Guerrini R, Salvadori S, Rizzi A, Regoli D, Calo G. Neurobiology, pharmacology, and medicinal chemistry of neuropeptide S and its receptor. Med Res Rev. 2010;30:751–777. - PubMed
-
- Xu YL, Reinscheid RK, Huitron-Resendiz S, Clark SD, Wang Z, et al. Neuropeptide S: a neuropeptide promoting arousal and anxiolytic-like effects. Neuron. 2004;43:487–497. - PubMed
-
- Reinscheid RK, Xu YL, Okamura N, Zeng J, Chung S, et al. Pharmacological characterization of human and murine neuropeptide s receptor variants. J Pharmacol Exp Ther. 2005;315:1338–1345. - PubMed
-
- Xu YL, Gall CM, Jackson VR, Civelli O, Reinscheid RK. Distribution of neuropeptide S receptor mRNA and neurochemical characteristics of neuropeptide S-expressing neurons in the rat brain. J Comp Neurol. 2007;500:84–102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

