Pepitome: evaluating improved spectral library search for identification complementarity and quality assessment
- PMID: 22217208
- PMCID: PMC3292681
- DOI: 10.1021/pr200874e
Pepitome: evaluating improved spectral library search for identification complementarity and quality assessment
Abstract
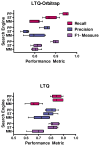
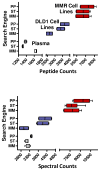
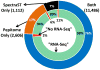
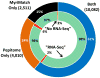
Spectral libraries have emerged as a viable alternative to protein sequence databases for peptide identification. These libraries contain previously detected peptide sequences and their corresponding tandem mass spectra (MS/MS). Search engines can then identify peptides by comparing experimental MS/MS scans to those in the library. Many of these algorithms employ the dot product score for measuring the quality of a spectrum-spectrum match (SSM). This scoring system does not offer a clear statistical interpretation and ignores fragment ion m/z discrepancies in the scoring. We developed a new spectral library search engine, Pepitome, which employs statistical systems for scoring SSMs. Pepitome outperformed the leading library search tool, SpectraST, when analyzing data sets acquired on three different mass spectrometry platforms. We characterized the reliability of spectral library searches by confirming shotgun proteomics identifications through RNA-Seq data. Applying spectral library and database searches on the same sample revealed their complementary nature. Pepitome identifications enabled the automation of quality analysis and quality control (QA/QC) for shotgun proteomics data acquisition pipelines.
Figures


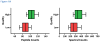
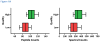
Similar articles
-
Spectral library generating function for assessing spectrum-spectrum match significance.J Proteome Res. 2013 Sep 6;12(9):3944-51. doi: 10.1021/pr400230p. Epub 2013 Jul 31. J Proteome Res. 2013. PMID: 23808827 Free PMC article.
-
ProMEX: a mass spectral reference database for proteins and protein phosphorylation sites.BMC Bioinformatics. 2007 Jun 23;8:216. doi: 10.1186/1471-2105-8-216. BMC Bioinformatics. 2007. PMID: 17587460 Free PMC article.
-
Spectrum-to-spectrum searching using a proteome-wide spectral library.Mol Cell Proteomics. 2011 Jul;10(7):M111.007666. doi: 10.1074/mcp.M111.007666. Epub 2011 Apr 30. Mol Cell Proteomics. 2011. PMID: 21532008 Free PMC article.
-
Building and searching tandem mass spectral libraries for peptide identification.Mol Cell Proteomics. 2011 Dec;10(12):R111.008565. doi: 10.1074/mcp.R111.008565. Epub 2011 Sep 6. Mol Cell Proteomics. 2011. PMID: 21900153 Free PMC article. Review.
-
Spectral library searching in proteomics.Proteomics. 2016 Mar;16(5):729-40. doi: 10.1002/pmic.201500296. Epub 2016 Feb 9. Proteomics. 2016. PMID: 26616598 Review.
Cited by
-
MS Ana: Improving Sensitivity in Peptide Identification with Spectral Library Search.J Proteome Res. 2023 Feb 3;22(2):462-470. doi: 10.1021/acs.jproteome.2c00658. Epub 2023 Jan 23. J Proteome Res. 2023. PMID: 36688604 Free PMC article.
-
Middle-down approach: a choice to sequence and characterize proteins/proteomes by mass spectrometry.RSC Adv. 2019 Jan 2;9(1):313-344. doi: 10.1039/c8ra07200k. eCollection 2018 Dec 19. RSC Adv. 2019. PMID: 35521579 Free PMC article. Review.
-
Chronic intermittent alcohol disrupts the GluN2B-associated proteome and specifically regulates group I mGlu receptor-dependent long-term depression.Addict Biol. 2017 Mar;22(2):275-290. doi: 10.1111/adb.12319. Epub 2015 Nov 8. Addict Biol. 2017. PMID: 26549202 Free PMC article.
-
Identification of Proteomic Features To Distinguish Benign Pulmonary Nodules from Lung Adenocarcinoma.J Proteome Res. 2017 Sep 1;16(9):3266-3276. doi: 10.1021/acs.jproteome.7b00245. Epub 2017 Aug 8. J Proteome Res. 2017. PMID: 28731711 Free PMC article.
-
Tracking the sources of blood meals of parasitic arthropods using shotgun proteomics and unidentified tandem mass spectral libraries.Nat Protoc. 2014 Apr;9(4):842-50. doi: 10.1038/nprot.2014.048. Epub 2014 Mar 13. Nat Protoc. 2014. PMID: 24625782 Free PMC article.
References
-
- Rodriguez H, Tezak Z, Mesri M, Carr SA, Liebler DC, Fisher SJ, Tempst P, Hiltke T, Kessler LG, Kinsinger CR, Philip R, Ransohoff DF, Skates SJ, Regnier FE, Anderson NL, Mansfield E. Analytical validation of protein-based multiplex assays: a workshop report by the NCI-FDA interagency oncology task force on molecular diagnostics. Clin Chem. 2010;56:237–243. - PubMed
-
- Lam H, Aebersold R. Using spectral libraries for peptide identification from tandem mass spectrometry (MS/MS) data. Curr Protoc Protein Sci. 2010;Chapter 25(Unit 25.5) - PubMed
-
- Deutsch EW. Tandem mass spectrometry spectral libraries and library searching. Methods Mol Biol. 2011;696:225–232. - PubMed
-
- Craig R, Cortens JC, Fenyo D, Beavis RC. Using annotated peptide mass spectrum libraries for protein identification. J Proteome Res. 2006;5:1843–1849. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

