Molecular determinants of enterovirus 71 viral entry: cleft around GLN-172 on VP1 protein interacts with variable region on scavenge receptor B 2
- PMID: 22219187
- PMCID: PMC3307280
- DOI: 10.1074/jbc.M111.301622
Molecular determinants of enterovirus 71 viral entry: cleft around GLN-172 on VP1 protein interacts with variable region on scavenge receptor B 2
Abstract
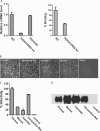
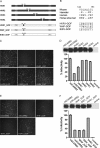
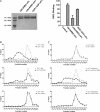
Enterovirus 71 (EV71) is one of the major pathogens that cause hand, foot, and mouth disease outbreaks in young children in the Asia-Pacific region in recent years. Human scavenger receptor class B 2 (SCARB2) is the main cellular receptor for EV71 on target cells. The requirements of the EV71-SCARB2 interaction have not been fully characterized, and it has not been determined whether SCARB2 serves as an uncoating receptor for EV71. Here we compared the efficiency of the receptor from different species including human, horseshoe bat, mouse, and hamster and demonstrated that the residues between 144 and 151 are critical for SCARB2 binding to viral capsid protein VP1 of EV71 and seven residues from the human receptor could convert murine SCARB2, an otherwise inefficient receptor, to an efficient receptor for EV71 viral infection. We also identified that EV71 binds to SCARB2 via a canyon of VP1 around residue Gln-172. Soluble SCARB2 could convert the EV71 virions from 160 S to 135 S particles, indicating that SCARB2 is an uncoating receptor of the virus. The uncoating efficiency of SCARB2 significantly increased in an acidic environment (pH 5.6). These studies elucidated the viral capsid and receptor determinants of enterovirus 71 infection and revealed a possible target for antiviral interventions.
Figures


References
-
- Racaniello V. R. (2007) in Fields Virology (Knipe D. M., Howley P. M., Griffin D. E., Lamb R. A., Martin M. A., Roizman B., Straus S. E., eds) 5 Ed., pp. 795–838, Vol. 1, Lippincott Williams & Wilkins, Philadelphia
-
- Huang C. C., Liu C. C., Chang Y. C., Chen C. Y., Wang S. T., Yeh T. F. (1999) Neurologic complications in children with enterovirus 71 infection. N. Engl. J. Med. 341, 936–942 - PubMed
-
- Lu M., Meng G., He Y. X., Zheng J., Liao S. L., Zhong Y. F., Zhao X. S., Shao H. Q., Wang Y. P., Gao Z. C., Gao Z. F. (2009) Pathology of enterovirus 71 infection: an autopsy study of 5 cases. Zhonghua Bing Li Xue Za Zhi 38, 81–85 - PubMed
-
- Lum L. C., Wong K. T., Lam S. K., Chua K. B., Goh A. Y. (1998) Neurogenic pulmonary oedema and enterovirus 71 encephalomyelitis. Lancet 352, 1391. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

