Afadin, a Ras/Rap effector that controls cadherin function, promotes spine and excitatory synapse density in the hippocampus
- PMID: 22219273
- PMCID: PMC3305287
- DOI: 10.1523/JNEUROSCI.4565-11.2012
Afadin, a Ras/Rap effector that controls cadherin function, promotes spine and excitatory synapse density in the hippocampus
Abstract
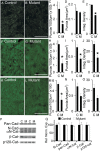
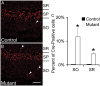
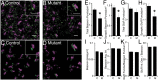
Many molecules regulate synaptogenesis, but intracellular signaling pathways required for their functions are poorly understood. Afadin is a Rap-regulated, actin-binding protein that promotes cadherin complex assembly as well as binding many other cell adhesion molecules and receptors. To examine its role in mediating synaptogenesis, we deleted afadin (mllt1), using a conditional allele, in postmitotic hippocampal neurons. Consistent with its role in promoting cadherin recruitment, afadin deletion resulted in 70% fewer and less intense N-cadherin puncta with similar reductions of β-catenin and αN-catenin puncta densities and 35% reduction in EphB2 puncta density. Its absence also resulted in 40% decreases in spine and excitatory synapse densities in the stratum radiatum of CA1, as determined by morphology, apposition of presynaptic and postsynaptic markers, and synaptic transmission. The remaining synapses appeared to function normally. Thus, afadin is a key intracellular signaling molecule for cadherin recruitment and is necessary for spine and synapse formation in vivo.
Figures







References
-
- Abe K, Chisaka O, Van Roy F, Takeichi M. Stability of dendritic spines and synaptic contacts is controlled by alpha N-catenin. Nat Neurosci. 2004;7:357–363. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
