The regulation of filamentous growth in yeast
- PMID: 22219507
- PMCID: PMC3249369
- DOI: 10.1534/genetics.111.127456
The regulation of filamentous growth in yeast
Abstract
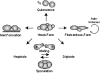
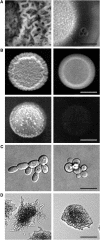
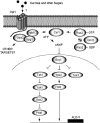
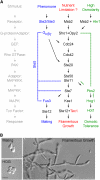
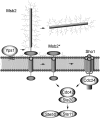
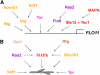
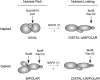
Filamentous growth is a nutrient-regulated growth response that occurs in many fungal species. In pathogens, filamentous growth is critical for host-cell attachment, invasion into tissues, and virulence. The budding yeast Saccharomyces cerevisiae undergoes filamentous growth, which provides a genetically tractable system to study the molecular basis of the response. Filamentous growth is regulated by evolutionarily conserved signaling pathways. One of these pathways is a mitogen activated protein kinase (MAPK) pathway. A remarkable feature of the filamentous growth MAPK pathway is that it is composed of factors that also function in other pathways. An intriguing challenge therefore has been to understand how pathways that share components establish and maintain their identity. Other canonical signaling pathways-rat sarcoma/protein kinase A (RAS/PKA), sucrose nonfermentable (SNF), and target of rapamycin (TOR)-also regulate filamentous growth, which raises the question of how signals from multiple pathways become integrated into a coordinated response. Together, these pathways regulate cell differentiation to the filamentous type, which is characterized by changes in cell adhesion, cell polarity, and cell shape. How these changes are accomplished is also discussed. High-throughput genomics approaches have recently uncovered new connections to filamentous growth regulation. These connections suggest that filamentous growth is a more complex and globally regulated behavior than is currently appreciated, which may help to pave the way for future investigations into this eukaryotic cell differentiation behavior.
Figures
References
-
- Arkinstall S. J., Papasavvas S. G., Payton M. A., 1991. Yeast alpha-mating factor receptor-linked G-protein signal transduction suppresses Ras-dependent activity. FEBS Lett. 284: 123–128 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

