A role for a single-stranded junction in RNA binding and specificity by the Tetrahymena group I ribozyme
- PMID: 22220837
- PMCID: PMC3277301
- DOI: 10.1021/ja2083575
A role for a single-stranded junction in RNA binding and specificity by the Tetrahymena group I ribozyme
Abstract
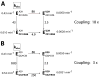
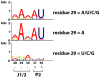
We have investigated the role of a single-stranded RNA junction, J1/2, that connects the substrate-containing P1 duplex to the remainder of the Tetrahymena group I ribozyme. Single-turnover kinetics, fluorescence anisotropy, and single-molecule fluorescence resonance energy transfer studies of a series of J1/2 mutants were used to probe the sequence dependence of the catalytic activity, the P1 dynamics, and the thermodynamics of docking of the P1 duplex into the ribozyme's catalytic core. We found that A29, the center A of three adenosine residues in J1/2, contributes 2 orders of magnitude to the overall ribozyme activity, and double-mutant cycles suggested that J1/2 stabilizes the docked state of P1 over the undocked state via a tertiary interaction involving A29 and the first base pair in helix P2 of the ribozyme, A31·U56. Comparative sequence analysis of this group I intron subclass suggests that the A29 interaction sets one end of a molecular ruler whose other end specifies the 5'-splice site and that this molecular ruler is conserved among a subclass of group I introns related to the Tetrahymena intron. Our results reveal substantial functional effects from a seemingly simple single-stranded RNA junction and suggest that junction sequences may evolve rapidly to provide important interactions in functional RNAs.
© 2012 American Chemical Society
Figures
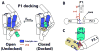

Similar articles
-
Characterization of a local folding event of the Tetrahymena group I ribozyme: effects of oligonucleotide substrate length, pH, and temperature on the two substrate binding steps.Biochemistry. 1999 Oct 26;38(43):14192-204. doi: 10.1021/bi9914309. Biochemistry. 1999. PMID: 10571993
-
Tertiary interactions with the internal guide sequence mediate docking of the P1 helix into the catalytic core of the Tetrahymena ribozyme.Biochemistry. 1993 Dec 14;32(49):13593-604. doi: 10.1021/bi00212a027. Biochemistry. 1993. PMID: 7504953
-
Evidence for processivity and two-step binding of the RNA substrate from studies of J1/2 mutants of the Tetrahymena ribozyme.Biochemistry. 1992 Feb 11;31(5):1386-99. doi: 10.1021/bi00120a015. Biochemistry. 1992. PMID: 1736996
-
A single-stranded junction modulates nanosecond motional ordering of the substrate recognition duplex of a group I ribozyme.Chembiochem. 2013 Sep 23;14(14):1720-3. doi: 10.1002/cbic.201300376. Epub 2013 Jul 30. Chembiochem. 2013. PMID: 23900919 Free PMC article.
-
The structure and function of catalytic RNAs.Sci China C Life Sci. 2009 Mar;52(3):232-44. doi: 10.1007/s11427-009-0038-z. Epub 2009 Mar 18. Sci China C Life Sci. 2009. PMID: 19294348 Review.
Cited by
-
Structural dynamics of a single-stranded RNA-helix junction using NMR.RNA. 2014 Jun;20(6):782-91. doi: 10.1261/rna.043711.113. Epub 2014 Apr 17. RNA. 2014. PMID: 24742933 Free PMC article.
-
Trans-splicing with the group I intron ribozyme from Azoarcus.RNA. 2014 Feb;20(2):202-13. doi: 10.1261/rna.041012.113. Epub 2013 Dec 16. RNA. 2014. PMID: 24344321 Free PMC article.
-
Mapping the kinetic barriers of a Large RNA molecule's folding landscape.PLoS One. 2014 Feb 25;9(2):e85041. doi: 10.1371/journal.pone.0085041. eCollection 2014. PLoS One. 2014. PMID: 24586236 Free PMC article.
-
Snapshots of the first-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.Nucleic Acids Res. 2023 Feb 22;51(3):1317-1325. doi: 10.1093/nar/gkac1268. Nucleic Acids Res. 2023. PMID: 36660826 Free PMC article.
-
Roles of long-range tertiary interactions in limiting dynamics of the Tetrahymena group I ribozyme.J Am Chem Soc. 2014 May 7;136(18):6643-8. doi: 10.1021/ja413033d. Epub 2014 Apr 28. J Am Chem Soc. 2014. PMID: 24738560 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

