A transcription activator-like effector toolbox for genome engineering
- PMID: 22222791
- PMCID: PMC3684555
- DOI: 10.1038/nprot.2011.431
A transcription activator-like effector toolbox for genome engineering
Abstract
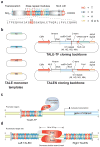
Transcription activator-like effectors (TALEs) are a class of naturally occurring DNA-binding proteins found in the plant pathogen Xanthomonas sp. The DNA-binding domain of each TALE consists of tandem 34-amino acid repeat modules that can be rearranged according to a simple cipher to target new DNA sequences. Customized TALEs can be used for a wide variety of genome engineering applications, including transcriptional modulation and genome editing. Here we describe a toolbox for rapid construction of custom TALE transcription factors (TALE-TFs) and nucleases (TALENs) using a hierarchical ligation procedure. This toolbox facilitates affordable and rapid construction of custom TALE-TFs and TALENs within 1 week and can be easily scaled up to construct TALEs for multiple targets in parallel. We also provide details for testing the activity in mammalian cells of custom TALE-TFs and TALENs using quantitative reverse-transcription PCR and Surveyor nuclease, respectively. The TALE toolbox described here will enable a broad range of biological applications.
Conflict of interest statement
The authors declare that they have no competing financial interests.
Figures





References
-
- Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. - PubMed
-
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. - PubMed
-
- Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

