Initiating RNA polymerase II and TIPs as hallmarks of enhancer activity and tissue-specificity
- PMID: 22223044
- PMCID: PMC3265787
- DOI: 10.4161/trns.2.6.18747
Initiating RNA polymerase II and TIPs as hallmarks of enhancer activity and tissue-specificity
Abstract
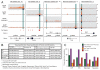
In past years, many efforts were invested to define epigenetic features associated with enhancers of transcription. We propose that both transcription initiation and the H3K4me3 histone modification are among the best hallmarks of active enhancers in several primary tissues and extend the concept of large transcription initiation platforms (TIPs).
© 2011 Landes Bioscience
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
