Compensatory functions of histone deacetylase 1 (HDAC1) and HDAC2 regulate transcription and apoptosis during mouse oocyte development
- PMID: 22223663
- PMCID: PMC3286984
- DOI: 10.1073/pnas.1118403109
Compensatory functions of histone deacetylase 1 (HDAC1) and HDAC2 regulate transcription and apoptosis during mouse oocyte development
Abstract
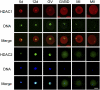
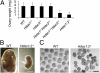
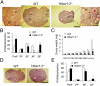
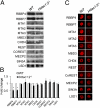
Dramatic changes in chromatin structure and histone modification occur during oocyte growth, as well as a global cessation of transcription. The role of histone modifications in these processes is poorly understood. We report the effect of conditionally deleting Hdac1 and Hdac2 on oocyte development. Deleting either gene has little or no effect on oocyte development, whereas deleting both genes results in follicle development arrest at the secondary follicle stage. This developmental arrest is accompanied by substantial perturbation of the transcriptome and a global reduction in transcription even though histone acetylation is markedly increased. There is no apparent change in histone repressive marks, but there is a pronounced decrease in histone H3K4 methylation, an activating mark. The decrease in H3K4 methylation is likely a result of increased expression of Kdm5b because RNAi-mediated targeting of Kdm5b in double-mutant oocytes results in an increase in H3K4 methylation. An increase in TRP53 acetylation also occurs in mutant oocytes and may contribute to the observed increased incidence of apoptosis. Taken together, these results suggest seminal roles of acetylation of histone and nonhistone proteins in oocyte development.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
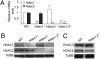


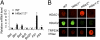

References
-
- Rodrigues P, Limback D, McGinnis LK, Plancha CE, Albertini DF. Oogenesis: Prospects and challenges for the future. J Cell Physiol. 2008;216:355–365. - PubMed
-
- van den Hurk R, Zhao J. Formation of mammalian oocytes and their growth, differentiation and maturation within ovarian follicles. Theriogenology. 2005;63:1717–1751. - PubMed
-
- Moore GP, Lintern-Moore S. Transcription of the mouse oocyte genome. Biol Reprod. 1978;18:865–870. - PubMed
-
- De La Fuente R, Eppig JJ. Transcriptional activity of the mouse oocyte genome: Companion granulosa cells modulate transcription and chromatin remodeling. Dev Biol. 2001;229:224–236. - PubMed
-
- Kageyama S, et al. Alterations in epigenetic modifications during oocyte growth in mice. Reproduction. 2007;133:85–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

