Regulation of DNA replication during development
- PMID: 22223677
- PMCID: PMC3252349
- DOI: 10.1242/dev.061838
Regulation of DNA replication during development
Abstract
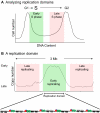
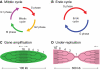
As development unfolds, DNA replication is not only coordinated with cell proliferation, but is regulated uniquely in specific cell types and organs. This differential regulation of DNA synthesis requires crosstalk between DNA replication and differentiation. This dynamic aspect of DNA replication is highlighted by the finding that the distribution of replication origins varies between differentiated cell types and changes with differentiation. Moreover, differential DNA replication in some cell types can lead to increases or decreases in gene copy number along chromosomes. This review highlights the recent advances and technologies that have provided us with new insights into the developmental regulation of DNA replication.
Figures

References
-
- Aggarwal B. D., Calvi B. R. (2004). Chromatin regulates origin activity in Drosophila follicle cells. Nature 430, 372–376 - PubMed
-
- Albertson D. G. (2006). Gene amplification in cancer. Trends Genet. 22, 447–455 - PubMed
-
- Araki H. (2010). Cyclin-dependent kinase-dependent initiation of chromosomal DNA replication. Curr. Opin. Cell Biol. 22, 766–771 - PubMed
-
- Arias E. E., Walter J. C. (2007). Strength in numbers: preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev. 21, 497–518 - PubMed
-
- Bell S. P., Dutta A. (2002). DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71, 333–374 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

