Origins and evolution of microRNA genes in plant species
- PMID: 22223755
- PMCID: PMC3318440
- DOI: 10.1093/gbe/evs002
Origins and evolution of microRNA genes in plant species
Abstract
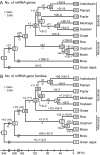
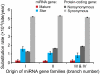
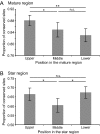
MicroRNAs (miRNAs) are among the most important regulatory elements of gene expression in animals and plants. However, their origin and evolutionary dynamics have not been studied systematically. In this paper, we identified putative miRNA genes in 11 plant species using the bioinformatic technique and examined their evolutionary changes. Our homology search indicated that no miRNA gene is currently shared between green algae and land plants. The number of miRNA genes has increased substantially in the land plant lineage, but after the divergence of eudicots and monocots, the number has changed in a lineage-specific manner. We found that miRNA genes have originated mainly by duplication of preexisting miRNA genes or protein-coding genes. Transposable elements also seem to have contributed to the generation of species-specific miRNA genes. The relative importance of these mechanisms in plants is quite different from that in Drosophila species, where the formation of hairpin structures in the genomes seems to be a major source of miRNA genes. This difference in the origin of miRNA genes between plants and Drosophila may be explained by the difference in the binding to target mRNAs between plants and animals. We also found that young miRNA genes are less conserved than old genes in plants as well as in Drosophila species. Yet, nearly half of the gene families in the ancestor of flowering plants have been lost in at least one species examined. This indicates that the repertoires of miRNA genes have changed more dynamically than previously thought during plant evolution.
Figures



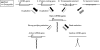
References
-
- Allen E, et al. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–1290. - PubMed
-
- Axtell MJ. Evolution of microRNAs and their targets: are all microRNAs biologically relevant? Biochim Biophys Acta. 2008;1779:725–734. - PubMed
-
- Axtell MJ, Bowman JL. Evolution of plant microRNAs and their targets. Trends Plant Sci. 2008;13:343–349. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

