BDDCS class prediction for new molecular entities
- PMID: 22224483
- PMCID: PMC3295927
- DOI: 10.1021/mp2004302
BDDCS class prediction for new molecular entities
Abstract
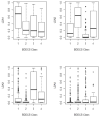
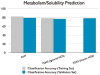
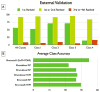
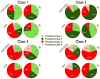
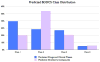
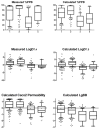
The Biopharmaceutics Drug Disposition Classification System (BDDCS) was successfully employed for predicting drug-drug interactions (DDIs) with respect to drug metabolizing enzymes (DMEs), drug transporters and their interplay. The major assumption of BDDCS is that the extent of metabolism (EoM) predicts high versus low intestinal permeability rate, and vice versa, at least when uptake transporters or paracellular transport is not involved. We recently published a collection of over 900 marketed drugs classified for BDDCS. We suggest that a reliable model for predicting BDDCS class, integrated with in vitro assays, could anticipate disposition and potential DDIs of new molecular entities (NMEs). Here we describe a computational procedure for predicting BDDCS class from molecular structures. The model was trained on a set of 300 oral drugs, and validated on an external set of 379 oral drugs, using 17 descriptors calculated or derived from the VolSurf+ software. For each molecule, a probability of BDDCS class membership was given, based on predicted EoM, FDA solubility (FDAS) and their confidence scores. The accuracy in predicting FDAS was 78% in training and 77% in validation, while for EoM prediction the accuracy was 82% in training and 79% in external validation. The actual BDDCS class corresponded to the highest ranked calculated class for 55% of the validation molecules, and it was within the top two ranked more than 92% of the time. The unbalanced stratification of the data set did not affect the prediction, which showed highest accuracy in predicting classes 2 and 3 with respect to the most populated class 1. For class 4 drugs a general lack of predictability was observed. A linear discriminant analysis (LDA) confirming the degree of accuracy for the prediction of the different BDDCS classes is tied to the structure of the data set. This model could routinely be used in early drug discovery to prioritize in vitro tests for NMEs (e.g., affinity to transporters, intestinal metabolism, intestinal absorption and plasma protein binding). We further applied the BDDCS prediction model on a large set of medicinal chemistry compounds (over 30,000 chemicals). Based on this application, we suggest that solubility, and not permeability, is the major difference between NMEs and drugs. We anticipate that the forecast of BDDCS categories in early drug discovery may lead to a significant R&D cost reduction.
Figures

Similar articles
-
Prediction of Biopharmaceutical Drug Disposition Classification System (BDDCS) by Structural Parameters.J Pharm Pharm Sci. 2019;22(1):247-269. doi: 10.18433/jpps30271. J Pharm Pharm Sci. 2019. PMID: 31287788
-
Provisional classification and in silico study of biopharmaceutical system based on caco-2 cell permeability and dose number.Mol Pharm. 2013 Jun 3;10(6):2445-61. doi: 10.1021/mp4000585. Epub 2013 May 15. Mol Pharm. 2013. PMID: 23675957
-
pH-Dependent solubility and permeability criteria for provisional biopharmaceutics classification (BCS and BDDCS) in early drug discovery.Mol Pharm. 2012 May 7;9(5):1199-212. doi: 10.1021/mp2004912. Epub 2012 Apr 26. Mol Pharm. 2012. PMID: 22489626
-
Predicting drug disposition via application of a Biopharmaceutics Drug Disposition Classification System.Basic Clin Pharmacol Toxicol. 2010 Mar;106(3):162-7. doi: 10.1111/j.1742-7843.2009.00498.x. Epub 2009 Dec 7. Basic Clin Pharmacol Toxicol. 2010. PMID: 20002064 Free PMC article. Review.
-
State of the Art and Uses for the Biopharmaceutics Drug Disposition Classification System (BDDCS): New Additions, Revisions, and Citation References.AAPS J. 2022 Feb 23;24(2):37. doi: 10.1208/s12248-022-00687-0. AAPS J. 2022. PMID: 35199251 Free PMC article. Review.
Cited by
-
Drug Disposition Classification Systems in Discovery and Development: A Comparative Review of the BDDCS, ECCS and ECCCS Concepts.Pharm Res. 2016 Nov;33(11):2583-93. doi: 10.1007/s11095-016-2001-6. Epub 2016 Jul 20. Pharm Res. 2016. PMID: 27439505 Review.
-
Neural Network Models for Predicting Solubility and Metabolism Class of Drugs in the Biopharmaceutics Drug Disposition Classification System (BDDCS).Eur J Drug Metab Pharmacokinet. 2024 Jan;49(1):1-6. doi: 10.1007/s13318-023-00861-5. Epub 2023 Oct 21. Eur J Drug Metab Pharmacokinet. 2024. PMID: 37864650
-
Can BDDCS illuminate targets in drug design?Drug Discov Today. 2019 Dec;24(12):2299-2306. doi: 10.1016/j.drudis.2019.09.021. Epub 2019 Oct 1. Drug Discov Today. 2019. PMID: 31585170 Free PMC article. Review.
-
Evaluation of the Relevance of DILI Predictive Hypotheses in Early Drug Development: Review of In Vitro Methodologies vs BDDCS Classification.Toxicol Res (Camb). 2018 May 8;7(3):358-370. doi: 10.1039/C8TX00016F. Epub 2018 Apr 18. Toxicol Res (Camb). 2018. PMID: 29785262 Free PMC article.
-
Dedication to professor Leslie Z. Benet: 50 years of scientific excellence and still going strong!Pharm Res. 2012 Sep;29(9):2345-53. doi: 10.1007/s11095-012-0808-3. Epub 2012 Jul 20. Pharm Res. 2012. PMID: 22814901 No abstract available.
References
-
- Amidon GL, Lennernas H, Shah VP, Crison JR. A theoretical basis for a biopharmaceutic drug classification: The correlation of in vitro drug product dissolution and in vivo bioavailability. Pharm Res. 1995;12:413–420. - PubMed
-
- FDA Guidance for Industry. Food and Drug Administration; Rockville, MD: 2000. Waiver of in vivo bioavailability and bioequivalence studies for immediate-release solid oral dosage forms based on a biopharmaceutics classification system.
-
- Wu CY, Benet LZ. Predicting drug disposition via application of BCS: transport/absorption/elimination interplay and development of a biopharmaceutics drug disposition classification system. Pharm Res. 2005;22:11–23. - PubMed
-
- Guideline on the investigation of bioequivalence. Committee for Medicinal Products for Human Use (CHMP) European Medicines Agency; London, UK: 2010.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

