Prognostic value of proliferation assay in the luminal, HER2-positive, and triple-negative biologic classes of breast cancer
- PMID: 22225836
- PMCID: PMC3496118
- DOI: 10.1186/bcr3084
Prognostic value of proliferation assay in the luminal, HER2-positive, and triple-negative biologic classes of breast cancer
Abstract
Introduction: Although the prognostic significance of proliferation in early invasive breast cancer has been recognized for a long time, recent gene-expression profiling studies have reemphasized its biologic and prognostic value and the potential application of its assessment in routine practice, particularly to define prognostic subgroups of luminal/hormone receptor-positive (HR+) tumors. This study aimed to assess the prognostic value of a proliferation assay by using Ki-67 immunohistochemistry as compared with mitotic count scores.
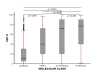
Method: Proliferation was assessed by using Ki-67 labeling index (Ki-67LI) and mitotic scores in a large (n = 1,550) and well-characterized series of clinically annotated primary operable invasive breast cancer with long-term follow-up. Tumors were phenotyped based on their IHC profiles into luminal/HR+, HER2+, and triple-negative (TN) classes. We used a split-sample development and validation approach to determine the optimal Ki-67LI cut-offs.
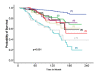
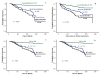
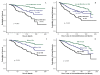
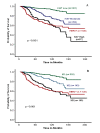
Results: The optimal cut-points of Ki-67LI were 10% and 50% for the luminal class. Both Ki7LI and MS were able to split luminal tumors into subgroups with significantly variable outcomes, independent of other variables. Neither mitotic count scores nor Ki-67LI was associated with outcome in the HER2+ or the TN classes.
Conclusions: Assessment of proliferation by using Ki-67LI and MS can distinguish subgroups of patients within luminal/hormone receptor-positive breast cancer significantly different in clinical outcomes. Overall, both Ki-67 LI and mitotic-count scores showed comparable results. The method described could provide a cost-effective method for prognostic subclassification of luminal/hormone receptor-positive breast cancer in routine clinical practice.
Figures
Similar articles
-
Validity of the proliferation markers Ki67, TOP2A, and RacGAP1 in molecular subgroups of breast cancer.Breast Cancer Res Treat. 2013 Jan;137(1):57-67. doi: 10.1007/s10549-012-2296-x. Epub 2012 Nov 8. Breast Cancer Res Treat. 2013. PMID: 23135572
-
Prognostic value of Ki-67 labeling index in patients with node-negative, triple-negative breast cancer.Breast Cancer Res Treat. 2012 Jul;134(1):277-82. doi: 10.1007/s10549-012-2040-6. Epub 2012 Apr 1. Breast Cancer Res Treat. 2012. PMID: 22467243
-
Power of PgR expression as a prognostic factor for ER-positive/HER2-negative breast cancer patients at intermediate risk classified by the Ki67 labeling index.BMC Cancer. 2017 May 22;17(1):354. doi: 10.1186/s12885-017-3331-4. BMC Cancer. 2017. PMID: 28532429 Free PMC article.
-
Prognostic Significance of Progesterone Receptor Expression in Estrogen-Receptor Positive, HER2-Negative, Node-Negative Invasive Breast Cancer With a Low Ki-67 Labeling Index.Clin Breast Cancer. 2017 Feb;17(1):41-47. doi: 10.1016/j.clbc.2016.06.012. Epub 2016 Jun 25. Clin Breast Cancer. 2017. PMID: 27477822
-
HER2 as a prognostic factor in breast cancer.Oncology. 2001;61 Suppl 2:67-72. doi: 10.1159/000055404. Oncology. 2001. PMID: 11694790 Review.
Cited by
-
Mediator complex (MED) 7: a biomarker associated with good prognosis in invasive breast cancer, especially ER+ luminal subtypes.Br J Cancer. 2018 Apr;118(8):1142-1151. doi: 10.1038/s41416-018-0041-x. Epub 2018 Mar 28. Br J Cancer. 2018. PMID: 29588513 Free PMC article.
-
Luminal breast cancer classification according to proliferative indices: clinicopathological characteristics and short-term survival analysis.Med Oncol. 2014 Jul;31(7):55. doi: 10.1007/s12032-014-0055-z. Epub 2014 Jun 17. Med Oncol. 2014. PMID: 24935624
-
P53 and Ki-67 as prognostic markers in triple-negative breast cancer patients.PLoS One. 2017 Feb 24;12(2):e0172324. doi: 10.1371/journal.pone.0172324. eCollection 2017. PLoS One. 2017. PMID: 28235003 Free PMC article.
-
MYC functions are specific in biological subtypes of breast cancer and confers resistance to endocrine therapy in luminal tumours.Br J Cancer. 2016 Apr 12;114(8):917-28. doi: 10.1038/bjc.2016.46. Epub 2016 Mar 8. Br J Cancer. 2016. PMID: 26954716 Free PMC article.
-
Prognostic and predictive value of Ki-67 in triple-negative breast cancer.Oncotarget. 2016 May 24;7(21):31079-87. doi: 10.18632/oncotarget.9075. Oncotarget. 2016. PMID: 27145269 Free PMC article.
References
-
- Daidone MG, Silvestrini R. Prognostic and predictive role of proliferation indices in adjuvant therapy of breast cancer. J Natl Cancer Inst Monogr. 2001;30:27–35. - PubMed
-
- Wirapati P, Sotiriou C, Kunkel S, Farmer P, Pradervand S, Haibe-Kains B, Desmedt C, Ignatiadis M, Sengstag T, Schütz F, Goldstein DR, Piccart M, Delorenzi M. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65. doi: 10.1186/bcr2124. - DOI - PMC - PubMed
-
- Weigelt B, Baehner FL, Reis-Filho JS. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J Pathol. 2010;220:263–80. - PubMed
-
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. - DOI - PubMed
-
- Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, Desmedt C, Larsimont D, Cardoso F, Peterse H, Nuyten D, Buyse M, Van de Vijver MJ, Bergh J, Piccart M, Delorenzi M. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–72. doi: 10.1093/jnci/djj052. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

