Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs
- PMID: 22227116
- PMCID: PMC3278580
- DOI: 10.1016/j.molcel.2011.10.023
Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs
Abstract
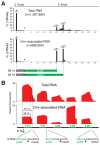
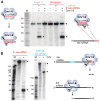
Small RNAs target invaders for silencing in the CRISPR-Cas pathways that protect bacteria and archaea from viruses and plasmids. The CRISPR RNAs (crRNAs) contain sequence elements acquired from invaders that guide CRISPR-associated (Cas) proteins back to the complementary invading DNA or RNA. Here, we have analyzed essential features of the crRNAs associated with the Cas RAMP module (Cmr) effector complex, which cleaves targeted RNAs. We show that Cmr crRNAs contain an 8 nucleotide 5' sequence tag (also found on crRNAs associated with other CRISPR-Cas pathways) that is critical for crRNA function and can be used to engineer crRNAs that direct cleavage of novel targets. We also present data that indicate that the Cmr complex cleaves an endogenous complementary RNA in Pyrococcus furiosus, providing direct in vivo evidence of RNA targeting by the CRISPR-Cas system. Our findings indicate that the CRISPR RNA-Cmr protein pathway may be exploited to cleave RNAs of interest.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Prokaryotic RNAi.Nat Methods. 2012 Mar;9(3):220-1. doi: 10.1038/nmeth.1916. Nat Methods. 2012. PMID: 22479705 No abstract available.
References
-
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. - PubMed
-
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

