Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae)
- PMID: 22228301
- PMCID: PMC3268322
- DOI: 10.1073/pnas.1112041109
Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae)
Abstract
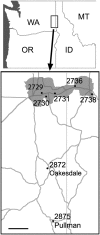
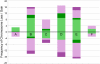
Polyploidy, or whole genome duplication, has played a major role in the evolution of many eukaryotic lineages. Although the prevalence of polyploidy in plants is well documented, the molecular and cytological consequences are understood largely from newly formed polyploids (neopolyploids) that have been grown experimentally. Classical cytological and molecular cytogenetic studies both have shown that experimental neoallopolyploids often have meiotic irregularities, producing chromosomally variable gametes and progeny; however, little is known about the extent or duration of chromosomal variation in natural neoallopolyploid populations. We report the results of a molecular cytogenetic study on natural populations of a neoallopolyploid, Tragopogon miscellus, which formed multiple times in the past 80 y. Using genomic and fluorescence in situ hybridization, we uncovered massive and repeated patterns of chromosomal variation in all populations. No population was fixed for a particular karyotype; 76% of the individuals showed intergenomic translocations, and 69% were aneuploid for one or more chromosomes. Importantly, 85% of plants exhibiting aneuploidy still had the expected chromosome number, mostly through reciprocal monosomy-trisomy of homeologous chromosomes (1:3 copies) or nullisomy-tetrasomy (0:4 copies). The extensive chromosomal variation still present after ca. 40 generations in this biennial species suggests that substantial and prolonged chromosomal instability might be common in natural populations after whole genome duplication. A protracted period of genome instability in neoallopolyploids may increase opportunities for alterations to genome structure, losses of coding and noncoding DNA, and changes in gene expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

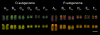

References
-
- Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–732. - PubMed
-
- Jiao Y, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. - PubMed
-
- Soltis DE, et al. Polyploidy and angiosperm diversification. Am J Bot. 2009;96:336–348. - PubMed
-
- De Bodt S, Maere S, Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evol. 2005;20:591–597. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

