Translation elongation regulates substrate selection by the signal recognition particle
- PMID: 22228766
- PMCID: PMC3293578
- DOI: 10.1074/jbc.M111.325001
Translation elongation regulates substrate selection by the signal recognition particle
Abstract
The signal recognition particle (SRP) is a universally conserved cellular machinery responsible for delivering membrane and secretory proteins to the proper cellular destination. The precise mechanism by which fidelity is achieved by the SRP pathway within the in vivo environment is yet to be understood. Previous studies have focused on the SRP pathway in isolation. Here we describe another important factor that modulates substrate selection by the SRP pathway: the ongoing synthesis of the nascent polypeptide chain by the ribosome. A slower translation elongation rate rescues the targeting defect of substrate proteins bearing mutant, suboptimal signal sequences both in vitro and in vivo. Consistent with a kinetic origin of this effect, similar rescue of protein targeting was also observed with mutant SRP receptors or SRP RNAs that specifically compromise the kinetics of SRP-receptor interaction during protein targeting. These data are consistent with a model in which ongoing protein translation is in constant kinetic competition with the targeting of the nascent proteins by the SRP and provides an important factor to regulate the fidelity of substrate selection by the SRP.
Figures
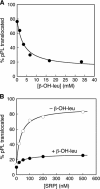

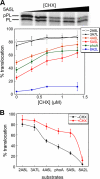
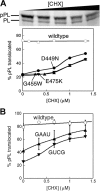

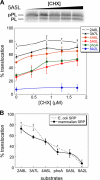
Similar articles
-
Signal recognition particle binds to translating ribosomes before emergence of a signal anchor sequence.Nucleic Acids Res. 2017 Nov 16;45(20):11858-11866. doi: 10.1093/nar/gkx888. Nucleic Acids Res. 2017. PMID: 29149347 Free PMC article.
-
SRP samples nascent chains for the presence of signal sequences by interacting with ribosomes at a discrete step during translation elongation.Cell. 1995 Jun 30;81(7):1075-84. doi: 10.1016/s0092-8674(05)80012-1. Cell. 1995. PMID: 7600575
-
Signal Recognition Particle Suppressor Screening Reveals the Regulation of Membrane Protein Targeting by the Translation Rate.mBio. 2021 Jan 12;12(1):e02373-20. doi: 10.1128/mBio.02373-20. mBio. 2021. PMID: 33436432 Free PMC article.
-
Dynamics of co-translational protein targeting.Curr Opin Chem Biol. 2015 Dec;29:79-86. doi: 10.1016/j.cbpa.2015.09.016. Epub 2015 Oct 30. Curr Opin Chem Biol. 2015. PMID: 26517565 Free PMC article. Review.
-
Fidelity of cotranslational protein targeting by the signal recognition particle.Annu Rev Biophys. 2014;43:381-408. doi: 10.1146/annurev-biophys-051013-022653. Annu Rev Biophys. 2014. PMID: 24895856 Free PMC article. Review.
Cited by
-
Mitochondrial mRNA localization is governed by translation kinetics and spatial transport.PLoS Comput Biol. 2022 Aug 19;18(8):e1010413. doi: 10.1371/journal.pcbi.1010413. eCollection 2022 Aug. PLoS Comput Biol. 2022. PMID: 35984860 Free PMC article.
-
Genomic differences between cultivated soybean, G. max and its wild relative G. soja.BMC Genomics. 2013;14 Suppl 1(Suppl 1):S5. doi: 10.1186/1471-2164-14-S1-S5. Epub 2013 Jan 21. BMC Genomics. 2013. PMID: 23368680 Free PMC article.
-
Local slowdown of translation by nonoptimal codons promotes nascent-chain recognition by SRP in vivo.Nat Struct Mol Biol. 2014 Dec;21(12):1100-5. doi: 10.1038/nsmb.2919. Epub 2014 Nov 24. Nat Struct Mol Biol. 2014. PMID: 25420103 Free PMC article.
-
Signal recognition particle-ribosome binding is sensitive to nascent chain length.J Biol Chem. 2014 Jul 11;289(28):19294-305. doi: 10.1074/jbc.M114.563239. Epub 2014 May 7. J Biol Chem. 2014. PMID: 24808175 Free PMC article.
-
Compensating Complete Loss of Signal Recognition Particle During Co-translational Protein Targeting by the Translation Speed and Accuracy.Front Microbiol. 2021 Jul 9;12:690286. doi: 10.3389/fmicb.2021.690286. eCollection 2021. Front Microbiol. 2021. PMID: 34305852 Free PMC article.
References
-
- Pool M. R. (2005) Signal recognition particles in chloroplasts, bacteria, yeast and mammals (review). Mol. Membr. Biol. 22, 3–15 - PubMed
-
- Walter P., Johnson A. E. (1994) Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 10, 87–119 - PubMed
-
- Walter P., Blobel G. (1983) Disassembly and reconstitution of signal recognition particle. Cell 34, 525–533 - PubMed
-
- Keenan R. J., Freymann D. M., Walter P., Stroud R. M. (1998) Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 94, 181–191 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

