Redox control of 20S proteasome gating
- PMID: 22229461
- PMCID: PMC3324812
- DOI: 10.1089/ars.2011.4210
Redox control of 20S proteasome gating
Abstract
The proteasome is the primary contributor in intracellular proteolysis. Oxidized or unstructured proteins can be degraded via a ubiquitin- and ATP-independent process by the free 20S proteasome (20SPT). The mechanism by which these proteins enter the catalytic chamber is not understood thus far, although the 20SPT gating conformation is considered to be an important barrier to allowing proteins free entrance. We have previously shown that S-glutathiolation of the 20SPT is a post-translational modification affecting the proteasomal activities.
Aims: The goal of this work was to investigate the mechanism that regulates 20SPT activity, which includes the identification of the Cys residues prone to S-glutathiolation.
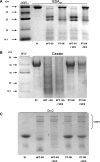
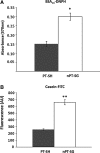
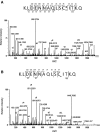
Results: Modulation of 20SPT activity by proteasome gating is at least partially due to the S-glutathiolation of specific Cys residues. The gate was open when the 20SPT was S-glutathiolated, whereas following treatment with high concentrations of dithiothreitol, the gate was closed. S-glutathiolated 20SPT was more effective at degrading both oxidized and partially unfolded proteins than its reduced form. Only 2 out of 28 Cys were observed to be S-glutathiolated in the proteasomal α5 subunit of yeast cells grown to the stationary phase in glucose-containing medium.
Innovation: We demonstrate a redox post-translational regulatory mechanism controlling 20SPT activity.
Conclusion: S-glutathiolation is a post-translational modification that triggers gate opening and thereby activates the proteolytic activities of free 20SPT. This process appears to be an important regulatory mechanism to intensify the removal of oxidized or unstructured proteins in stressful situations by a process independent of ubiquitination and ATP consumption. Antioxid. Redox Signal. 16, 1183-1194.
Figures


Similar articles
-
Mutations of Cys and Ser residues in the α5-subunit of the 20S proteasome from Saccharomyces cerevisiae affects gating and chronological lifespan.Arch Biochem Biophys. 2019 May 15;666:63-72. doi: 10.1016/j.abb.2019.03.012. Epub 2019 Mar 30. Arch Biochem Biophys. 2019. PMID: 30940569
-
Redox regulation of the proteasome via S-glutathionylation.Redox Biol. 2013 Dec 14;2:44-51. doi: 10.1016/j.redox.2013.12.003. Redox Biol. 2013. PMID: 24396728 Free PMC article. Review.
-
20S proteasome activity is modified via S-glutathionylation based on intracellular redox status of the yeast Saccharomyces cerevisiae: implications for the degradation of oxidized proteins.Arch Biochem Biophys. 2014 Sep 1;557:65-71. doi: 10.1016/j.abb.2014.05.002. Epub 2014 May 9. Arch Biochem Biophys. 2014. PMID: 24813691
-
The physiological role of the free 20S proteasome in protein degradation: A critical review.Biochim Biophys Acta Gen Subj. 2018 Dec;1862(12):2948-2954. doi: 10.1016/j.bbagen.2018.09.009. Epub 2018 Sep 16. Biochim Biophys Acta Gen Subj. 2018. PMID: 30297324 Review.
-
Peptides that activate the 20S proteasome by gate opening increased oxidized protein removal and reduced protein aggregation.Free Radic Biol Med. 2014 Feb;67:304-13. doi: 10.1016/j.freeradbiomed.2013.11.017. Epub 2013 Nov 27. Free Radic Biol Med. 2014. PMID: 24291399
Cited by
-
Activators of the 26S proteasome when protein degradation increases.Exp Mol Med. 2025 Feb;57(1):41-49. doi: 10.1038/s12276-024-01385-x. Epub 2025 Jan 8. Exp Mol Med. 2025. PMID: 39779978 Free PMC article. Review.
-
The Cys Sense: Thiol Redox Switches Mediate Life Cycles of Cellular Proteins.Biomolecules. 2021 Mar 22;11(3):469. doi: 10.3390/biom11030469. Biomolecules. 2021. PMID: 33809923 Free PMC article. Review.
-
Early cysteine-dependent inactivation of 26S proteasomes does not involve particle disassembly.Redox Biol. 2018 Jun;16:123-128. doi: 10.1016/j.redox.2018.02.016. Epub 2018 Feb 22. Redox Biol. 2018. PMID: 29499565 Free PMC article.
-
Quantitative analysis of redox proteome reveals oxidation-sensitive protein thiols acting in fundamental processes of developmental hematopoiesis.Redox Biol. 2022 Jul;53:102343. doi: 10.1016/j.redox.2022.102343. Epub 2022 May 23. Redox Biol. 2022. PMID: 35640380 Free PMC article.
-
The Role and Regulation of Autophagy and the Proteasome During Aging and Senescence in Plants.Genes (Basel). 2019 Apr 2;10(4):267. doi: 10.3390/genes10040267. Genes (Basel). 2019. PMID: 30987024 Free PMC article. Review.
References
-
- Asher G. Reuven N. Shaul Y. 20S proteasomes and protein degradation “by default”. Bioessays. 2006;28:844–849. - PubMed
-
- Babbitt SE. Kiss A. Deffenbaugh AE. Chang YH. Bailly E. Erdjument-Bromage H. Tempst P. Buranda T. Sklar LA. Baumler J. Gogol E. Skowyra D. ATP hydrolysis-dependent disassembly of the 26S proteasome is part of the catalytic cycle. Cell. 2005;121:553–565. - PubMed
-
- Bajorek M. Finley D. Glickman MH. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr Biol. 2003;13:1140–1144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

