A novel µ-conopeptide, CnIIIC, exerts potent and preferential inhibition of NaV1.2/1.4 channels and blocks neuronal nicotinic acetylcholine receptors
- PMID: 22229737
- PMCID: PMC3419909
- DOI: 10.1111/j.1476-5381.2012.01837.x
A novel µ-conopeptide, CnIIIC, exerts potent and preferential inhibition of NaV1.2/1.4 channels and blocks neuronal nicotinic acetylcholine receptors
Abstract
Background and purpose: The µ-conopeptide family is defined by its ability to block voltage-gated sodium channels (VGSCs), a property that can be used for the development of myorelaxants and analgesics. We characterized the pharmacology of a new µ-conopeptide (µ-CnIIIC) on a range of preparations and molecular targets to assess its potential as a myorelaxant.
Experimental approach: µ-CnIIIC was sequenced, synthesized and characterized by its direct block of elicited twitch tension in mouse skeletal muscle and action potentials in mouse sciatic and pike olfactory nerves. µ-CnIIIC was also studied on HEK-293 cells expressing various rodent VGSCs and also on voltage-gated potassium channels and nicotinic acetylcholine receptors (nAChRs) to assess cross-interactions. Nuclear magnetic resonance (NMR) experiments were carried out for structural data.
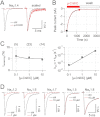
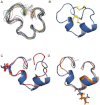
Key results: Synthetic µ-CnIIIC decreased twitch tension in mouse hemidiaphragms (IC(50) = 150 nM), and displayed a higher blocking effect in mouse extensor digitorum longus muscles (IC = 46 nM), compared with µ-SIIIA, µ-SmIIIA and µ-PIIIA. µ-CnIIIC blocked Na(V)1.4 (IC(50) = 1.3 nM) and Na(V)1.2 channels in a long-lasting manner. Cardiac Na(V)1.5 and DRG-specific Na(V)1.8 channels were not blocked at 1 µM. µ-CnIIIC also blocked the α3β2 nAChR subtype (IC(50) = 450 nM) and, to a lesser extent, on the α7 and α4β2 subtypes. Structure determination of µ-CnIIIC revealed some similarities to α-conotoxins acting on nAChRs.
Conclusion and implications: µ-CnIIIC potently blocked VGSCs in skeletal muscle and nerve, and hence is applicable to myorelaxation. Its atypical pharmacological profile suggests some common structural features between VGSCs and nAChR channels.
© 2012 The Authors. British Journal of Pharmacology © 2012 The British Pharmacological Society.
Figures





References
-
- Bax A, Davis DG. MLEV-17 based two-dimensional homonuclear magnetization transfer spectroscopy. J Magn Reson. 1985;65:355–360.
-
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
-
- Bulaj G, West PJ, Garrett JE, Watkins M, Zhang MM, Norton RS, et al. Novel conotoxins from Conus striatus and Conus kinoshitai selectively block TTX-resistant sodium channels. Biochemistry. 2005;44:7259–7265. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

