On the molecular mechanism of GC content variation among eubacterial genomes
- PMID: 22230424
- PMCID: PMC3274465
- DOI: 10.1186/1745-6150-7-2
On the molecular mechanism of GC content variation among eubacterial genomes
Abstract
Background: As a key parameter of genome sequence variation, the GC content of bacterial genomes has been investigated for over half a century, and many hypotheses have been put forward to explain this GC content variation and its relationship to other fundamental processes. Previously, we classified eubacteria into dnaE-based groups (the dimeric combination of DNA polymerase III alpha subunits), according to a hypothesis where GC content variation is essentially governed by genome replication and DNA repair mechanisms. Further investigation led to the discovery that two major mutator genes, polC and dnaE2, may be responsible for genomic GC content variation. Consequently, an in-depth analysis was conducted to evaluate various potential intrinsic and extrinsic factors in association with GC content variation among eubacterial genomes.
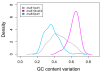
Results: Mutator genes, especially those with dominant effects on the mutation spectra, are biased towards either GC or AT richness, and they alter genomic GC content in the two opposite directions. Increased bacterial genome size (or gene number) appears to rely on increased genomic GC content; however, it is unclear whether the changes are directly related to certain environmental pressures. Certain environmental and bacteriological features are related to GC content variation, but their trends are more obvious when analyzed under the dnaE-based grouping scheme. Most terrestrial, plant-associated, and nitrogen-fixing bacteria are members of the dnaE1|dnaE2 group, whereas most pathogenic or symbiotic bacteria in insects, and those dwelling in aquatic environments, are largely members of the dnaE1|polV group.
Conclusion: Our studies provide several lines of evidence indicating that DNA polymerase III α subunit and its isoforms participating in either replication (such as polC) or SOS mutagenesis/translesion synthesis (such as dnaE2), play dominant roles in determining GC variability. Other environmental or bacteriological factors, such as genome size, temperature, oxygen requirement, and habitat, either play subsidiary roles or rely indirectly on different mutator genes to fine-tune the GC content. These results provide a comprehensive insight into mechanisms of GC content variation and the robustness of eubacterial genomes in adapting their ever-changing environments over billions of years.
Figures




References
-
- Li W, Grauer D. Fundamentals of Molecular Evolution. First. Sunderland MA: Sinauer Associates Inc; 1991.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

