Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon
- PMID: 22231304
- PMCID: PMC3260867
- DOI: 10.1084/jem.20101387
Inflammation switches the differentiation program of Ly6Chi monocytes from antiinflammatory macrophages to inflammatory dendritic cells in the colon
Abstract
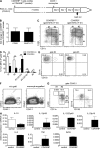
Dendritic cells (DCs) and macrophages (MPs) are important for immunological homeostasis in the colon. We found that F4/80(hi)CX3CR1(hi) (CD11b(+)CD103(-)) cells account for 80% of mouse colonic lamina propria MHC-II(hi) cells. Both CD11c(+) and CD11c(-) cells within this population were identified as MPs based on multiple criteria, including an MP transcriptome revealed by microarray analysis. These MPs constitutively released high levels of IL-10 at least partially in response to the microbiota via an MyD88-independent mechanism. In contrast, cells expressing low to intermediate levels of F4/80 and CX3CR1 were identified as DCs based on phenotypic and functional analysis and comprise three separate CD11c(hi) cell populations: CD103(+)CX3CR1(-)CD11b(-) DCs, CD103(+)CX3CR1(-)CD11b(+) DCs, and CD103(-)CX3CR1(int)CD11b(+) DCs. In noninflammatory conditions, Ly6C(hi) monocytes (MOs) differentiated primarily into CD11c(+) but not CD11c(-) MPs. In contrast, during colitis, Ly6C(hi) MOs massively invaded the colon and differentiated into proinflammatory CD103(-)CX3CR1(int)CD11b(+) DCs, which produced high levels of IL-12, IL-23, iNOS, and TNF. These findings demonstrate the dual capacity of Ly6C(hi) blood MOs to differentiate into either regulatory MPs or inflammatory DCs in the colon and that the balance of these immunologically antagonistic cell types is dictated by microenvironmental conditions.
Figures



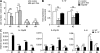


References
-
- Cheong C., Matos I., Choi J.H., Dandamudi D.B., Shrestha E., Longhi M.P., Jeffrey K.L., Anthony R.M., Kluger C., Nchinda G., et al. 2010. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell. 143:416–429 10.1016/j.cell.2010.09.039 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

