Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel
- PMID: 22231484
- PMCID: PMC3267885
- DOI: 10.1038/ng.1042
Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel
Abstract
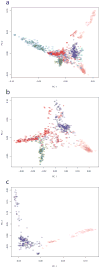
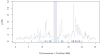
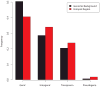
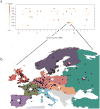
Arabidopsis thaliana is native to Eurasia and is naturalized across the world. Its ability to be easily propagated and its high phenotypic variability make it an ideal model system for functional, ecological and evolutionary genetics. To date, analyses of the natural genetic variation of A. thaliana have involved small numbers of individual plants or genetic markers. Here we genotype 1,307 worldwide accessions, including several regional samples, using a 250K SNP chip. This allowed us to produce a high-resolution description of the global pattern of genetic variation. We applied three complementary selection tests and identified new targets of selection. Further, we characterized the pattern of historical recombination in A. thaliana and observed an enrichment of hotspots in its intergenic regions and repetitive DNA, which is consistent with the pattern that is observed for humans but which is strikingly different from that observed in other plant species. We have made the seeds we used to produce this Regional Mapping (RegMap) panel publicly available. This panel comprises one of the largest genomic mapping resources currently available for global natural isolates of a non-human species.
Figures

Comment in
-
Arabidopsis thaliana as a model for the genetics of local adaptation.Nat Genet. 2012 Jan 27;44(2):115-6. doi: 10.1038/ng.1079. Nat Genet. 2012. PMID: 22281764
References
-
- Kim S, et al. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet. 2007;39:1151–5. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

